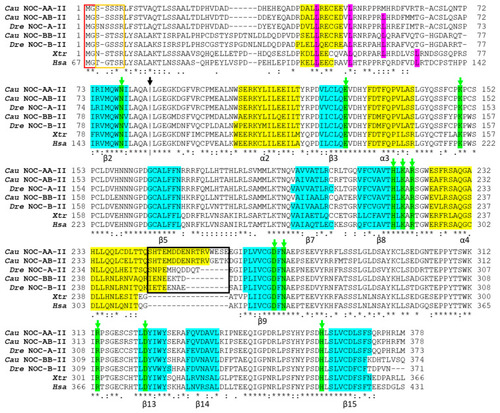
Figure 7
- ID
- ZDB-FIG-240113-16
- Publication
- Madera et al., 2023 - Gene Characterization of Nocturnin Paralogues in Goldfish: Full Coding Sequences, Structure, Phylogeny and Tissue Expression
- Other Figures
- All Figure Page
- Back to All Figure Page
|
Alignment of the deduced amino acid sequences of splicing variant II of goldfish NOC-AA (Cau NOC-AA-II: WNX29031), NOC-AB (Cau NOC-AB-II: WNX29026), and NOC-BB (Cau NOC-BB-II: WNX29028) with NOC sequences from |