Fig. 7
- ID
- ZDB-FIG-231215-160
- Publication
- Gupta et al., 2022 - Coordinated activities of Myosin Vb isoforms and mTOR signaling regulate epithelial cell morphology during development
- Other Figures
- All Figure Page
- Back to All Figure Page
|
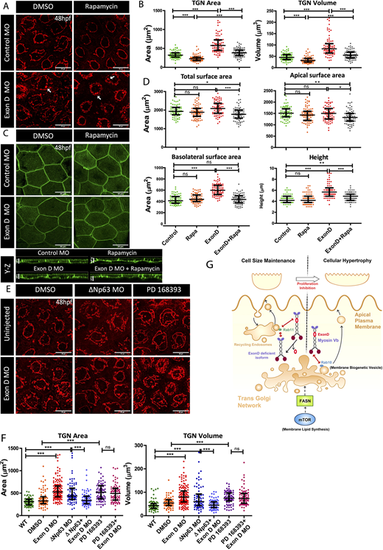
Myosin Vb and mTOR co-ordination at the TGN. (A,C) Confocal scans of the peridermal cells showing TGN (A) and cell membranes (C) of 48 hpf Tg(cldnB:lynEGFP) embryos injected with Exon D morpholino (Exon D MO) and the control embryos treated with DMSO or Rapamycin. (B,D) Quantification of the TGN surface area and volume (B) and total, apical and basolateral cell surface area and cell height (D) under different genetic conditions mentioned along the x-axes. (E,F) TGN morphologies (E) and quantifications of TGN surface area and volume (F) from the uninjected and Exon D morphant embryos either co-injected with ΔNp63 morpholino (ΔNp63 MO) or treated with PD16393 drug. (G) A schematic of Myosin Vb isoforms showing different outcomes depending upon their interaction with either Rab11 or Rab10. Although Myosin Vb-Rab11 interaction is essential to maintain the cell size during development, presumably by regulating recycling of the apical endosomes, MyosinVb-Rab 10 interaction regulates the apically directed transport from TGN downstream of the mTOR-FASN axis, during compensatory cell growth. Data are median±interquartile range. *P<0.05, **P<0.01, ***P<0.001 (Kruskal–Wallis test with Dunn's post hoc test). ns, not significant. Note that only essential pairwise comparisons are shown in the figure panels B,D and F. For the complete list of pairwise comparisons please refer to Table S6. Scale bars: 20 µm (X-Y plane of A,C,E); 5 µm (Y-Z plane of C). |