Fig. 4
|
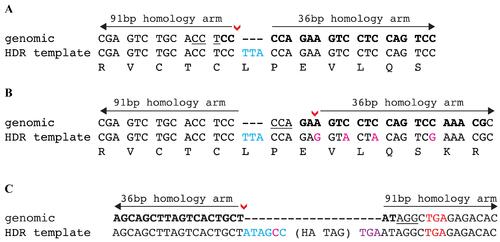
Examples of HDR template design to knock-in desired DNA sequences. (A) The DNA insertion of an in-frame codon TTA occurs directly at the DSB (denoted by red arrowhead). This insertion disrupts the sgRNA binding site and no additional modifications are required. (B) The DNA insertions occur 5 bp upstream of the DSB (denoted by red arrowhead). The insertion does not disrupt the sgRNA binding or the PAM site and a silent change cannot be made to the PAM site. Therefore, several silent changes (shown in magenta) are required to prevent recutting after HDR repair. (C) The HA epitope tag is inserted at the 3′ end of a gene’s ORF. The DSB (denoted by red arrowhead) occurs 6 bp upstream of the stop codon (shown in red font). To maintain the gene’s ORF, these 6 bp (shown in blue font) are added into the HDR template with one of the nucleotides (shown in magenta font) changed to a silent change to modify PAM site and prevent recutting after HDR repair. A new stop codon (shown in purple font) is added after the HA tag to terminate translation. In all examples, the sgRNA is in bold and the PAM site is underlined |