Fig. 2
- ID
- ZDB-FIG-220718-25
- Publication
- Darp et al., 2022 - Oncogenic BRAF induces whole-genome doubling through suppression of cytokinesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
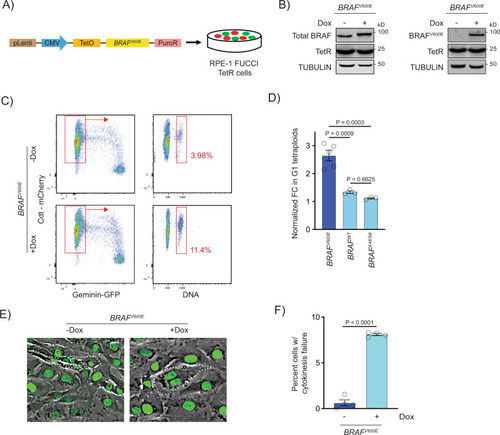
BRAFV600E-induced binucleate, tetraploid cells arise via cytokinesis failure.
A Generation of BRAFV600E-expressing RPE-1 FUCCI cell lines with a lentiviral-based doxycycline-inducible vector. B Western blot showing inducible expression of BRAFV600E (+Dox) in RPE-1 FUCCI cells using a BRAF and BRAFV600E-specific antibody. Expression of the Tet repressor protein is shown. TUBULIN was used as the loading control. A representative of three independent biological replicates is shown. C Flow cytometry plots of control (−Dox) and BRAFV600E-expressing (+Dox) RPE-1 FUCCI cells. Tetraploid cells accumulating in G1 were quantified based on Cdt1-mCherry positivity and Hoechst incorporation. Percentages of G1 tetraploid cells in control and BRAFV600E-expressing cultures are indicated. D Fold change (FC) in G1 tetraploids normalized to the control (−Dox) are shown for BRAFV600E, BRAFWT and BRAFK483W (kinase-dead) -expressing RPE-1 FUCCI cell lines. N = 5 independent experiments for BRAFV600E, N = 3 for BRAFWT and N = 3 for BRAFK483W. One-way ANOVA with Tukey’s multiple comparisons test. Error bars represent mean ± SEM. E Merged phase contrast and GFP photomicrographs of H2B-GFP expressing control (−Dox) and BRAFV600E-expressing (+Dox) RPE-1 cells that have recently undergone mitosis. White dotted lines indicate 2 cells with 1 nucleus each that have separated following a successful cytokinesis (−Dox) and 1 cell with 2 nuclei that has failed cytokinesis (+Dox). F Percentages of cells exhibiting cytokinesis failure in H2B-GFP RPE-1 control (−Dox) and BRAFV600E-expressing (+Dox) cells. N = 4 independent experiments examining -Dox = 934 and +Dox = 568 cells. Unpaired Student’s t test. Error bars represent mean ± SEM. |