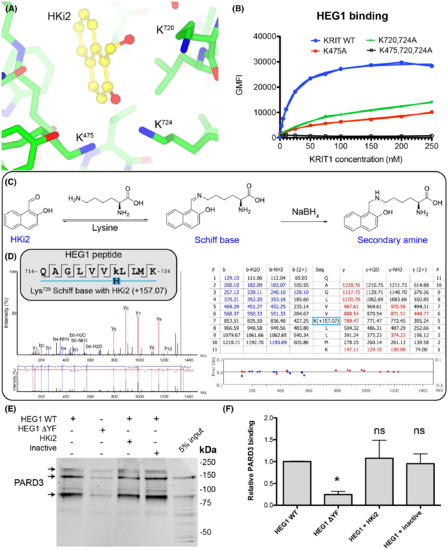
Fig. 5
|
KRIT1 Lys720 forms a covalent reversible bond with the aldehyde of HKi2 and HKi2 does not block PARD3 binding to HEG1. (A) KRIT1 bound to HKi2 highlighting the position of three lysines residues near the HKi2 aldehyde. HKi2 is shown in yellow and KRIT1 residues in green. (B) All tested EGFP-KRIT1 FERM domain mutants tested had reduced HEG1 binding. (C) Expected lysine modification upon treatment with sodium borohydryde to “trap” the Schiff base. (D) LC–MS/MS results. The searches in Peaks used a mass of 157.0653 (in monoisotopic mass) to lysine residues as variable modification. The tryptic peptide containing Lys720 showed such a mass for a lysine residue confirming that it forms a Schiff base with HKi2. (E) HUVEC lysates were incubated with either HEG1 WT or HEG1 ΔYF matrix and western blotted for PARD3. The mixture contained either DMSO, HKi2 or an inactive compound, 2-hydroxy-1-naphthoic acid (compound 9). The binding of PARD3 to HEG1 ΔYF is largely reduced in comparison to HEG1 WT, but neither HKi2 nor an inactive compound, 2-hydroxy-1-naphthoic acid, affected the binding. (F) Relative PARD3 binding from three independent experiments. Mean with SD are shown. ANOVA with a Tukey post hoc test: *, p < 0.05. |