|
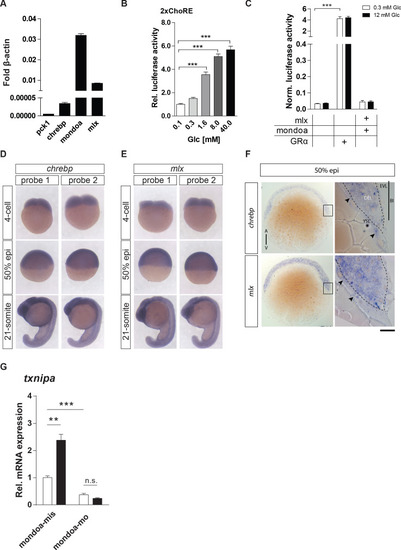
Additional data of Mondo pathway characterization experiments in zebrafish.(A) RT-qPCR determination of transcript levels of the Mondo pathway factors chrebp, mondoa and mlx in PAC2 cells shows that all three factors are expressed in this zebrafish cell line. The liver-specific gene phosphoenolpyruvate carboxykinase 1 (pck1) was used as a negative control (n = 3). (B) Reporter activity from the 2xChoRE reporter construct showed a significant dose-dependent increase upon glucose treatment in human HepG2 cells (n = 4). (C) Overexpression studies with mondoa and mlx in PAC2 cells with a GRE reporter construct. Cells were treated with either low (0.3 mM; white bars) or high (12 mM; black bars) glucose (Glc) concentrations and bioluminescence was measured after 24 hr of incubation (n = 4). Data were normalized to Renilla luciferase activity to control for transfection efficiency (Norm. bioluminescence). No glucose induction or regulation mediated by Mondo pathway factors was observed for the GRE reporter construct. (D, E) Analysis of spatial distribution of chrebp (D) and mlx (E) transcripts at 4 cell, 50% epiboly (50% epi) and 21-somite stages by WISH. For both genes, expression was detected in the blastodisc and is ubiquitous during later stages. The same staining pattern was observed for two probes directed against different portions of each transcript (probe 1 and probe 2). (F) Epon sections of 50% epiboly (50% epi.) stage embryos show chrebp (upper panels) and mlx (lower panels) expression in the enveloping layer (EVL) and in the deep cell layer (DEL) of the blastoderm (Bl) as well as in the yolk syncyctial layer (YSL, arrowheads). Scale bar: D, E, 0.36 mm; F, 0.13 mm for left panels, 20 µm for right panels. (G) Glucose induction of the MondoA target gene txnipa is MondoA dependent in zebrafish embryos. Embryos were injected with the glucose analog 2-deoxy-D-glucose (2-DG; black bars) or with water (white bars) as a control along with either mondoa-mis or mondoa-mo. RNA was extracted at the sphere stage to perform RT-qPCR of thioredoxin-interacting protein a (txnipa) (n = 3, 20 embryos each). Error bars represent SEM, *, p≤0.05; **, p≤0.01; ***, p≤0.001.
|