|
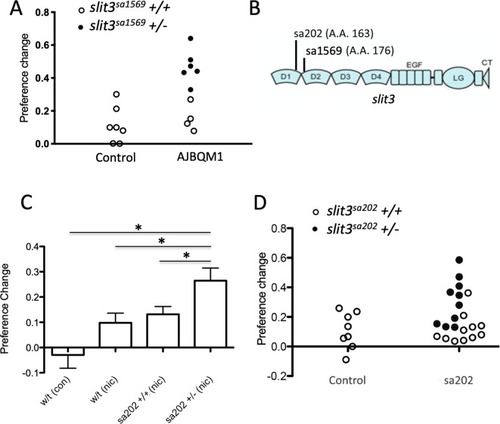
<italic>slit3</italic> mutations segregate with nicotine place preference.(A) Segregation of slit3sa1569 mutation with nicotine seeking. CPP change scores for individual un-mutagenized TLF wild type fish (n = 7) and AJBQM1 fish (n = 10). Following CPP analysis, fish were genotyped for 25 loss of function mutations contained within the family. Black dots indicate slit3sa1569/+ heterozygous mutant fish. White dots indicate slit3sa1569+/+ fish. Heterozygosity for slit3sa1569 segregates with increased nicotine seeking behaviour. (B) Position of ENU-induced mutations in zebrafish Slit3 protein. slit3sa1569 (A > G transition) disrupts a splice site in intron seven affecting translation at amino acid 176. slit3sa202 (G > T transversion) introduces a stop codon at amino acid 163. Both mutations truncate the protein before the leucine rich repeat domain 2 (D2), which interacts with membrane bound ROBO during SLIT-ROBO signalling. (C) Nicotine preference of slit3sa202 line. slit3sa202/+ fish (n = 18) show increased nicotine preference compared to wild type TLF controls (n = 8) (p=0.001) and wild type siblings slit3+/+ (n = 14) (p<0.05). Bars indicate mean + SEM. (D) Segregation of slit3sa202 allele with nicotine seeking. CPP change scores for individual un-mutagenised TLF wild type parent strain fish (n = 8) and slit3sa202 fish (n = 21). Black dots indicate slit3sa202/+ heterozygous mutant fish, white dots indicate slit3sa202+/+ fish. Mutations in slit3sa202 co-segregate with nicotine preference. Heterozygous slit3+/sa202 present increased place preference compared to slit3sa202+/+ siblings (n = 11).
|