Fig. S3
- ID
- ZDB-FIG-120412-38
- Publication
- Camp et al., 2012 - Intronic Cis-Regulatory Modules Mediate Tissue-Specific and Microbial Control of angptl4/fiaf Transcription
- Other Figures
- All Figure Page
- Back to All Figure Page
|
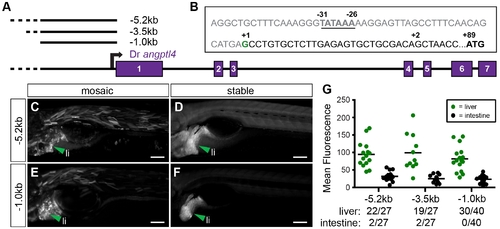
Non-coding DNA upstream of the zebrafish angptl4 transcription start site drives expression in the liver but not in the intestine or islet. (A) The zebrafish angptl4 locus and positions of promoter regions assayed in 0–7 dpf transgenic zebrafish are annotated to scale. (B) 52 RACE and EST data (not shown) establish a single transcription start site directly upstream of exon 1. The positions of the TATA box, transcription start site, and translation start site are annotated. (C, E) Non-coding DNA -5.2 kb and -1 kb upstream of the translation start site drives expression in the liver in 6 dpf mosaic animals. Note that the -5.2 kb fragment includes a region -4.9 kb upstream from the TSS that shares extensive homology with medaka (see Figure 2A). Scale bars = 50 μm. (D, F) Liver expression pattern is confirmed in the F1 generation of injected animals harboring stable insertions of the -5.2 kb (Tg(-5.2angptl4:GFP)) and -1 kb transgenes (Tg(-1angptl4:GFP)). Scale bars = 50 μm. (G) Fluorescence intensity in mosaic animals is quantified (see Materials and Methods) in the liver and intestine. Circles represent mean fluorescence averaged in three mosaic patches within the liver (green) or intestine (black) of 1 fish. Note that there is minimal to no reporter expression in either the intestine or the islet (not shown). Ratios of liver or intestine positive fish versus total fish expressing GFP are shown below the corresponding construct name. |