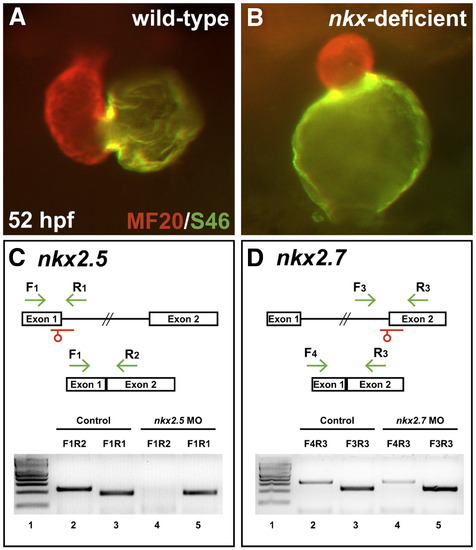
Splice MOs confirm specificity and efficacy of nkx2.5 and nkx2.7 knockdown. (A,B) Frontal views, dorsal to the top at 52 hpf. MF20/S46 immunofluorescence distinguishes ventricular myocardium (red) from atrial myocardium (yellow). Injection of anti-nkx2.5 and anti-nkx2.7 splice MOs has effects similar to that of injection of anti-nkx2.5 and anti-nkx2.7 ATG MOs (Fig. 1D). (C,D) MO injection impairs splicing of nkx2.5 and nkx2.7. Schematics depict the structures of the nkx2.5 (C) and nkx2.7 (D) pre-mRNA and mRNA. Locations of splice MOs are indicated in red. Primer pairs used for RT-PCR are indicated in green. Gels document RT-PCR amplification products, comparing detection of spliced (lanes 2 and 4) and unspliced (lanes 3 and 5) messages in cDNA from 26 hpf control embryos and embryos injected with anti-nkx2.5 splice MO or anti-nkx2.7 splice MO, respectively. Primer pair F1R1 amplifies 189 bp of unspliced nkx2.5 pre-mRNA, primer pair F1R2 amplifies 230 bp of spliced nkx2.5 mRNA, primer pair F3R3 amplifies 229 bp of unspliced nkx2.7 pre-mRNA, and primer pair F4R3 amplifies 323 bp of spliced nkx2.7 mRNA. In both gels, lane 1 contains a 100 bp molecular weight ladder (NEB, Ipswich, MA, USA). Embryos injected with anti-nkx2.5 splice MO exhibit a loss of spliced nkx2.5 mRNA, and embryos injected with anti-nkx2.7 splice MO demonstrate a partial reduction in spliced nkx2.7 mRNA with a concomitant increase in unspliced pre-mRNA. Note that the predicted PCR products with intronic inclusion for the unspliced nkx2.5 (1.4 kb) and nkx2.7 (2.2 kb) pre-mRNAs are absent, most likely because PCR conditions did not favor amplification of larger products.
|

