- Title
-
Pulsed stimulated Brillouin microscopy enables high-sensitivity mechanical imaging of live and fragile biological specimens
- Authors
- Yang, F., Bevilacqua, C., Hambura, S., Neves, A., Gopalan, A., Watanabe, K., Govendir, M., Bernabeu, M., Ellenberg, J., Diz-Muñoz, A., Köhler, S., Rapti, G., Jechlinger, M., Prevedel, R.
- Source
- Full text @ Nat. Methods
|
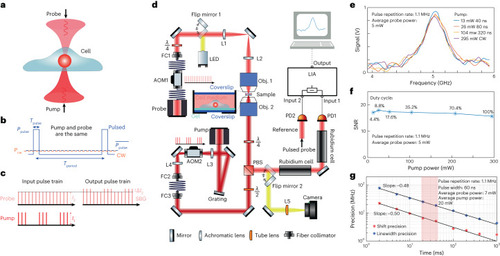
Pulsed-SBS approach and performance. |
|
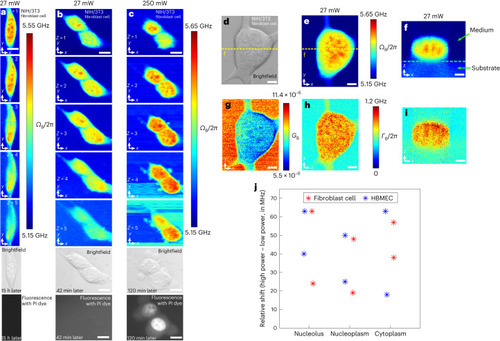
Pulsed-SBS imaging of cultured cells. |
|
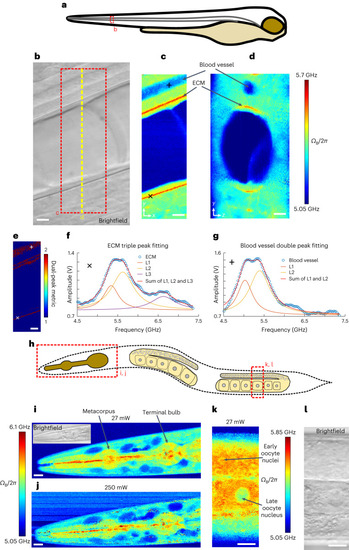
Pulsed-SBS imaging of zebrafish larvae and young adult |
|
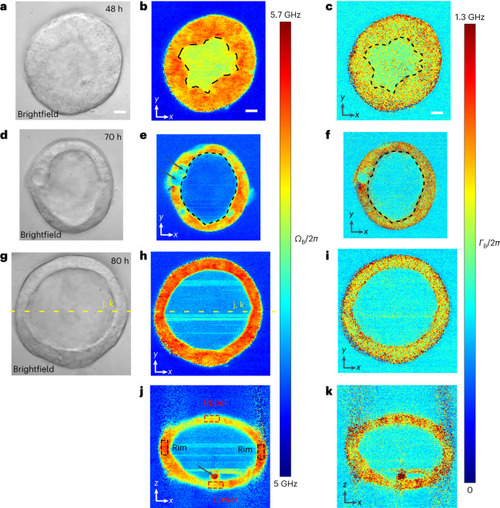
Pulsed-SBS imaging of mouse mammary gland organoids. |
|
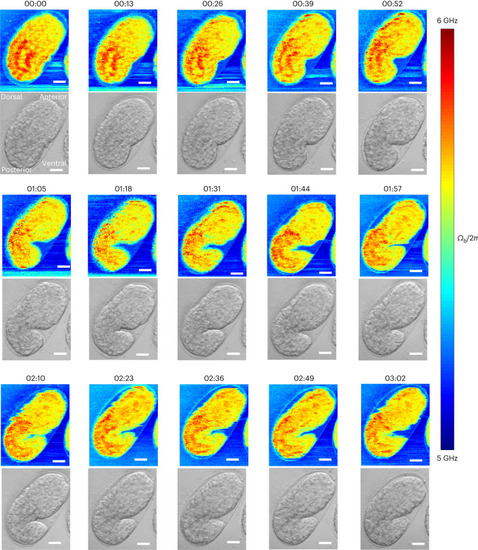
Time-lapse pulsed-SBS imaging of Brillouin shift (top) and brightfield (bottom) images over a 3-h time span and at 13-min time interval (representative of |