- Title
-
Sulindac selectively induces autophagic apoptosis of GABAergic neurons and alters motor behaviour in zebrafish
- Authors
- Sun, W., Wang, M., Zhao, J., Zhao, S., Zhu, W., Wu, X., Li, F., Liu, W., Wang, Z., Gao, M., Zhang, Y., Xu, J., Zhang, M., Wang, Q., Wen, Z., Shen, J., Zhang, W., Huang, Z.
- Source
- Full text @ Nat. Commun.
|
Sulindac, identified through a small clinical anti-inflammatory drug screening, elicited neurodevelopmental toxicity. |
|
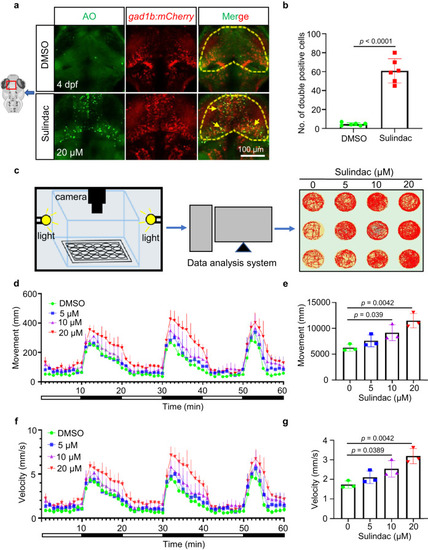
Sulindac selectively induced apoptosis of GABAergic neurons and altered motor behaviour in zebrafish larvae. |
|
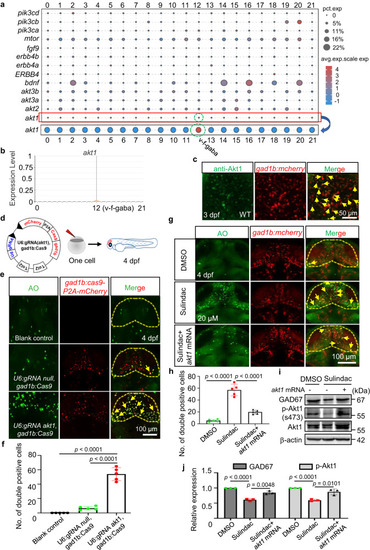
Sulindac selectively induced apoptosis of GABAergic neurons via Akt signalling in zebrafish. |
|
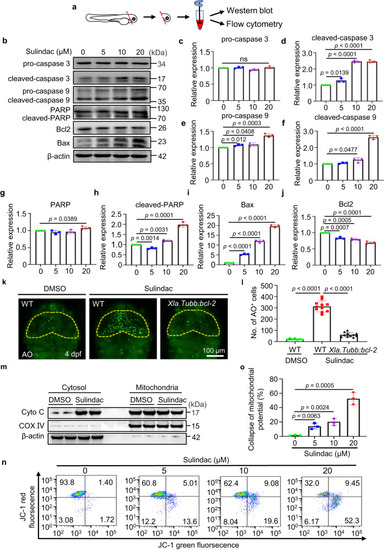
Sulindac induces apoptosis via the mitochondrial pathway. |
|
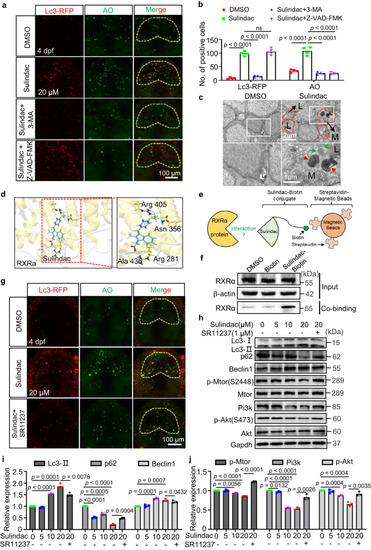
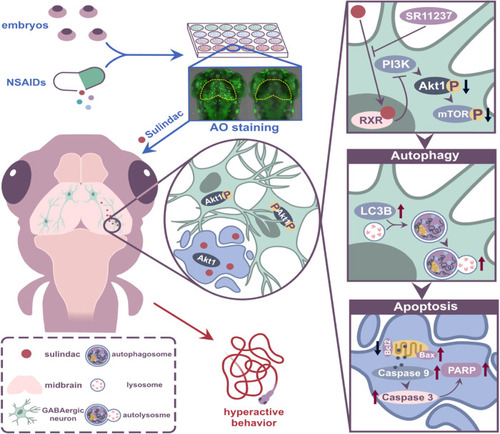
Sulindac promoted RXRa-dependent autophagy-induced apoptosis through the PI3K/AKT/mTOR pathway in zebrafish neurons. |
|
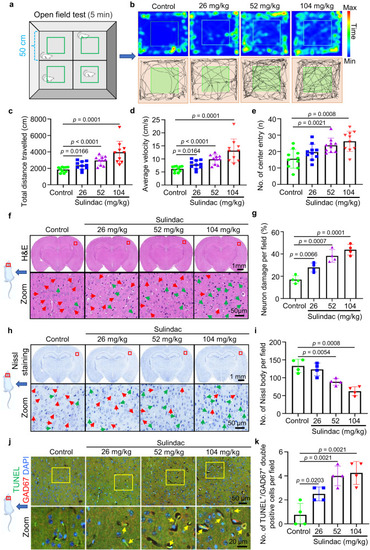
Sulindac caused mice to display hyperactivity and induced apoptosis of neocortical GABAergic neurons. |
|
Schematic illustration of the proposed mechanisms. Sulindac treatment induces RXRa-dependent autophagic cell death in GABAergic neurons via the PI3K/AKT/mTOR pathway, leading to hyperactive behaviour in zebrafish larvae. |