- Title
-
Heterogeneity in quiescent Müller glia in the uninjured zebrafish retina drive differential responses following photoreceptor ablation
- Authors
- Krylov, A., Yu, S., Veen, K., Newton, A., Ye, A., Qin, H., He, J., Jusuf, P.R.
- Source
- Full text @ Front. Mol. Neurosci.
|
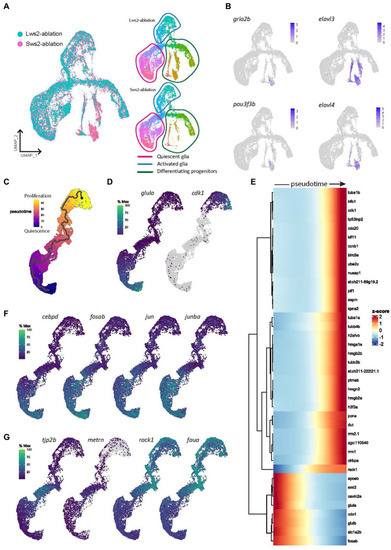
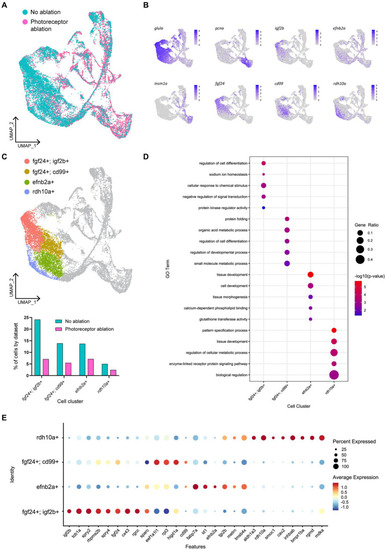
Gene expression modules in activated Müller glia following photoreceptor ablation. |
|
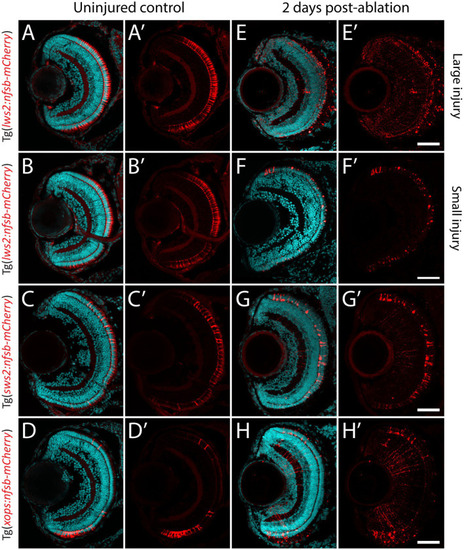
Photoreceptor ablation paradigms differing in subtype targeted, injury extent and injury location. |
|
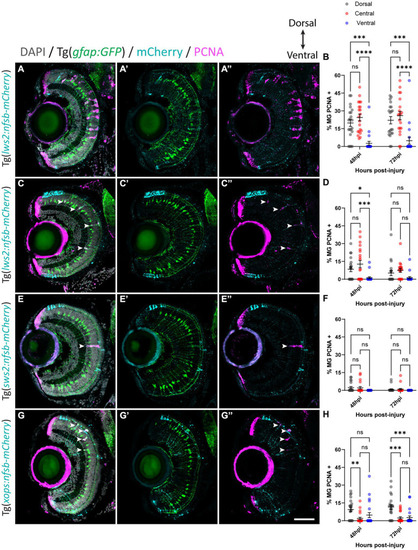
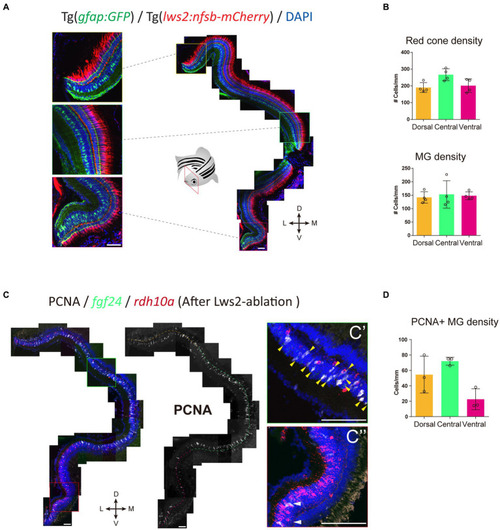
Müller glia subpopulations along the dorsal to ventral axis differ in their regenerative ability. Response of Müller glia following widespread Lws2 |
|
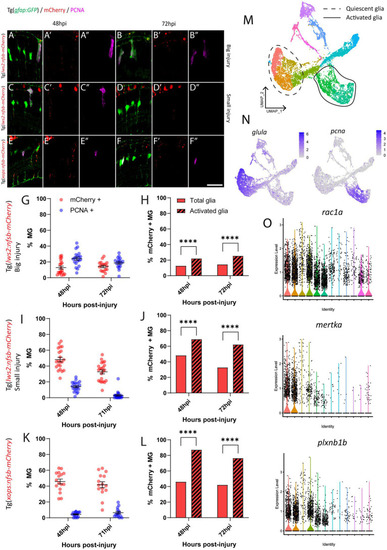
Investigation of phagocytosis and proliferation by Müller glia following photoreceptor ablation. |
|
Heterogeneity exists in Müller glia of the uninjured zebrafish retina. |
|
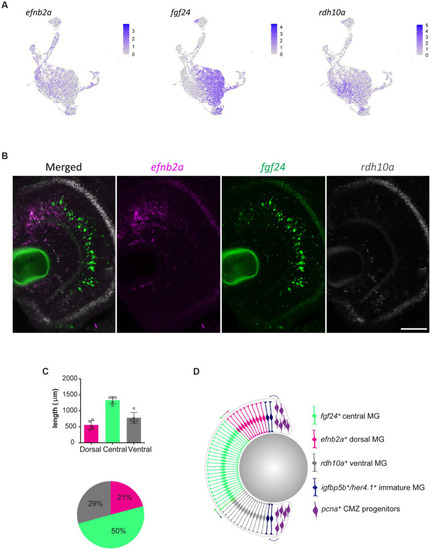
Molecularly distinct Müller glia subpopulations differ in their spatial location. |
|
|
|
Müller glia heterogeneity persists in the presence of photoreceptor ablation. |