- Title
-
Nerve growth factor receptor (Ngfr) induces neurogenic plasticity by suppressing reactive astroglial Lcn2/Slc22a17 signaling in Alzheimer's disease
- Authors
- Siddiqui, T., Cosacak, M.I., Popova, S., Bhattarai, P., Yilmaz, E., Lee, A.J., Min, Y., Wang, X., Allen, M., İş, Ö., Atasavum, Z.T., Rodriguez-Muela, N., Vardarajan, B.N., Flaherty, D., Teich, A.F., Santa-Maria, I., Freudenberg, U., Werner, C., Tosto, G., Mayeux, R., Ertekin-Taner, N., Kizil, C.
- Source
- Full text @ NPJ Regen Med
|
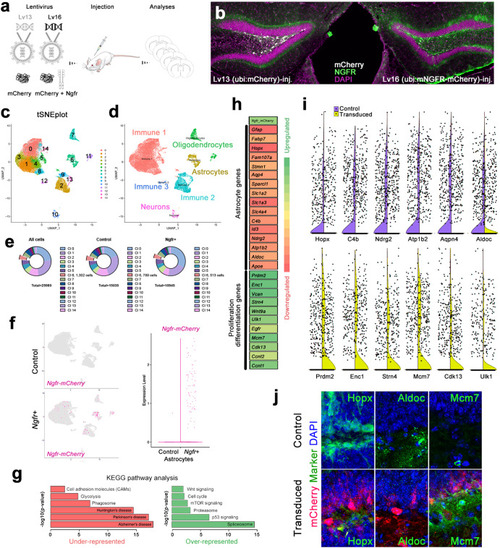
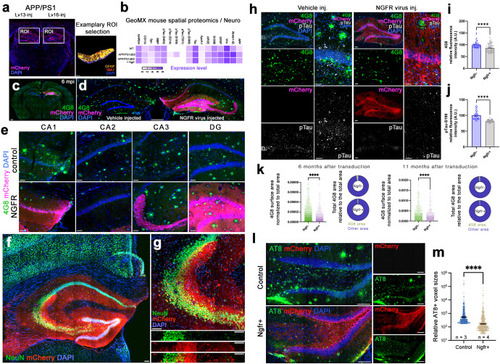
NGFR enhances proliferative and neurogenic markers in dentate gyrus (DG) astrocytes. |
|
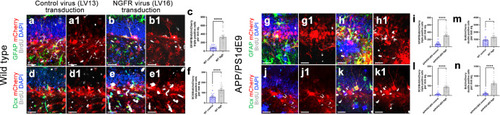
NGFR promotes proliferation of DG astrocytes and neurogenesis in wild type and APP/PS1dE9 model of AD. Immunostaining for GFAP, BrdU and mCherry with DAPI counterstain in Lv13- ( |
|
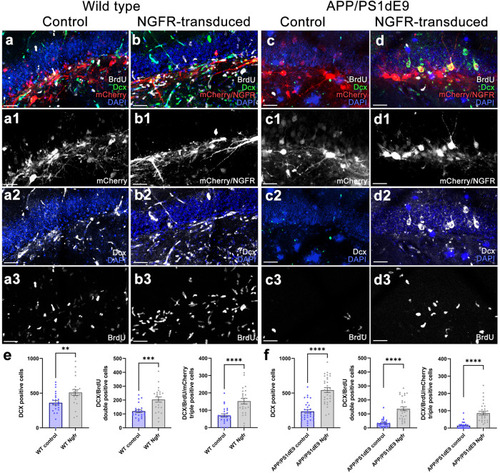
Lv16 transduction enhances neurogenesis in wild type and APP/PS1dE9 mouse model of Alzheimer’s disease. |
|
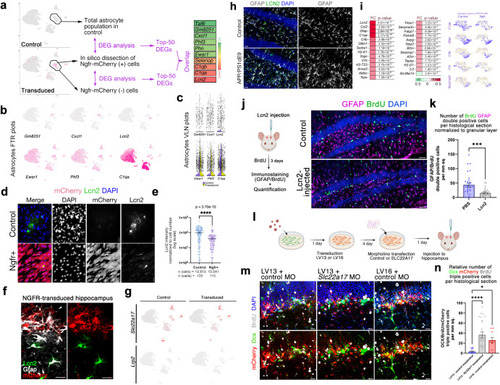
Ngfr regulates neurogenic response in DG astrocytes through suppression of Lcn2/Slc22a17 activity. |
|
Ngfr reduces Aβ42 load and phosphorylated Tau in the hippocampus of APP/PS1dE9 mice. |
|
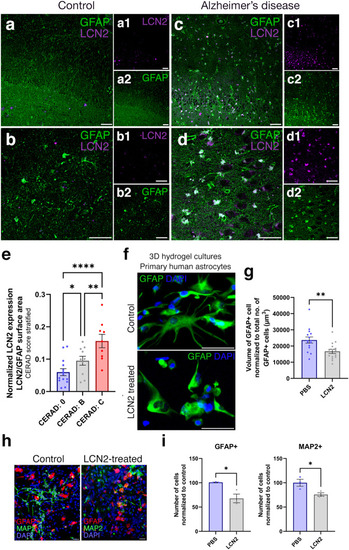
LCN2 is upregulated in human brains with AD. Immunohistochemical stainings for LCN2 and GFAP on hippocampal brain sections of healthy control ( |
|
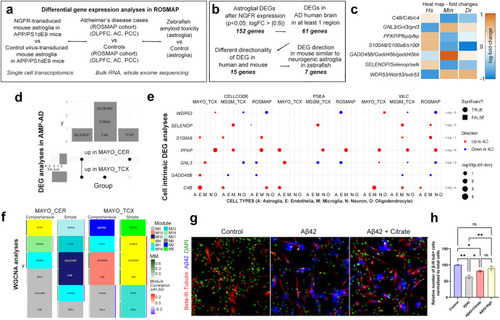
Comparison of gene expression changes in mouse brain with NGFR to human AD cohorts by differential gene expression analyses, cell intrinsic gene expression and Weighted gene co-expression network analysis. |
|
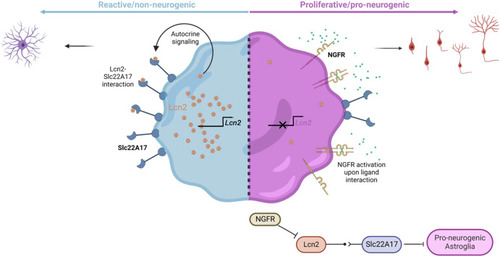
Schematic view of the NGFR/LCN2/SLC22A17-dependent neurogenic switch mechanism in astroglia. Nerve growth factor receptor can convert reactive astroglia to neurogenic state in mouse model of Alzheimer’s disease (AD) through suppression of Lcn2 activity on Slc22a17 receptor. In AD cohorts in humans, |