- Title
-
A New Transgenic Tool to Study the Ret Signaling Pathway in the Enteric Nervous System
- Authors
- Bandla, A., Melancon, E., Taylor, C.R., Davidson, A.E., Eisen, J.S., Ganz, J.
- Source
- Full text @ Int. J. Mol. Sci.
|
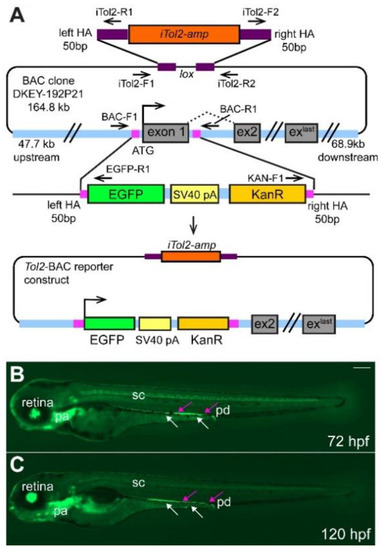
Overview of BAC-construct cloning strategy. (A) The start codon and exon 1 of ret were replaced by homologous recombination with the EGFP-SV40-pA-KanR construct using 50 bp-long homology arms (HA, magenta box) as indicated. iTol2 sites were included in the BAC backbone as shown using homologous recombination. The arrows indicate forward and reverse primer pairs to verify the correct generation of the tol2-BAC reporter construct. Integration of the BAC DNA does not result in overexpression of ret as the ret ATG is replaced by the EGFP cassette and the EGFP insert contains a strong transcription termination signal (SV40 pA, simian virus 40 poly A) [30]. Overview of GFP+ cells in the retina, pharyngeal arches (pa), spinal cord (sc), pronephric duct (pd, magenta arrows), and ENS cells (white arrows) at 72 (B) and 120 (C) hours post fertilization (hpf). (B,C): Whole-mount side-views of embryo/larva at the stage indicated. KanR kanamycin. Scale bar = 200 µm. |
|
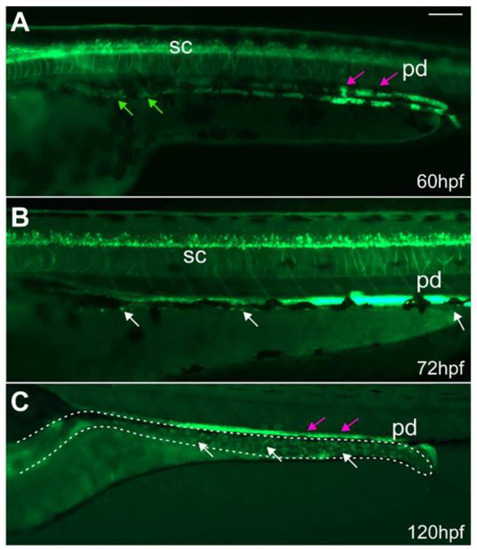
ret:GFP+ EPCs migrate along the developing gut and populate the gut. At (A) 60 hpf, GFP+ EPCs (green arrows) migrate along the developing gut. At (B) 72 and (C) 120 hpf, GFP+ ENS cells (white arrows) populate the gut (dashed line). Some of the GFP+ cells in the gut are enteroendocrine cells. The GFP+ pronephric ducts (pd, magenta arrows) directly overlay the ENS. (A–C): Whole-mount side-views of embryos/larvae at the stage indicated. Dashed line outlines the gut. Scale bar = 100 µm. |
|
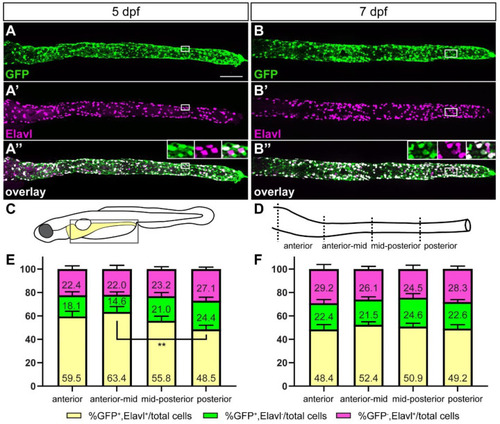
The majority of GFP+ cells are ENS neurons at later stages of ENS development. At 5 (A–A”) and 7 (B–B”) days post fertilization (dpf) GFP+ ENS neurons (white) are found along the whole length of the gut. A smaller fraction are GFP+ non-neuronal cells (green) or GFP− ENS neurons (magenta). Insets in (A”,B”) show the close-up of the boxed area, GFP, Elavl, and overlay from left to right. Schematic of boxed area of larval schematic (C) indicates the four gut sub-regions analyzed (D). Quantification of GFP and Elavl colocalization at 5 dpf (E) and 7 dpf (F) in four gut regions shown in (D). Bar graph shows % GFP+/Elavl+ out of total number of cells quantified [GFP+/Elavl+, GFP+/Elavl−, and GFP−/Elavl+], (yellow), % GFP+/Elavl− out of total cells (green), and % GFP−/Elavl+ out of total cells (magenta). Using 2-way ANOVA, we did not find significant differences between the cell populations of the different gut regions except where indicated (** p ≤ 0.01). Error bars show ±standard error of the mean (5 dpf: 2 experiments, 19 larvae; 7 dpf: 2 experiments, 15 larvae). (A–A”,B–B”): maximum projections of dissected guts at stage indicated. Scale bar = 100 µm. |
|
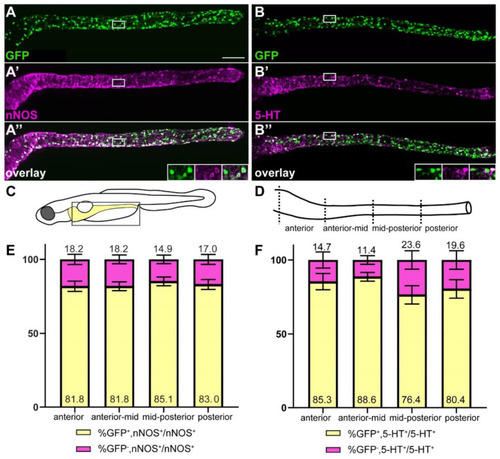
The majority of nitrergic and serotonergic neurons are GFP+. ret:GFP is expressed in the majority of nitrergic (A–A”) and serotonergic (B–B”) neurons (white). Insets in (A”,B”) show close-up of the boxed area, GFP, nNOS or 5-HT, and overlay from left to right. Schematic of boxed area of larval schematic (C) indicates the four gut sub-regions analyzed (D). (E) Quantification of GFP and nNOS colocalization in four gut regions as shown in (D). Bar graph shows % GFP+/nNOS+ out of total nNOS+ cells (yellow), and % GFP−/nNOS+ out of total nNOS+ cells (magenta). Error bars show ±standard error of the mean (2 experiments, 23 larvae). (F) Quantification of GFP and 5-HT colocalization in four gut regions shown in D. Bar graph shows % GFP+/5-HT+ out of total 5-HT+ cells (yellow), and % GFP−/5-HT+ out of total 5-HT+ cells (magenta). Using 2-way ANOVA, we did not find significant differences between the cell population of the different gut regions. Error bars show ±standard error of the mean (2 experiments, 22 larvae). (A–A”,B–B”): maximum projections of dissected guts. Scale bar = 100 µm. |
|
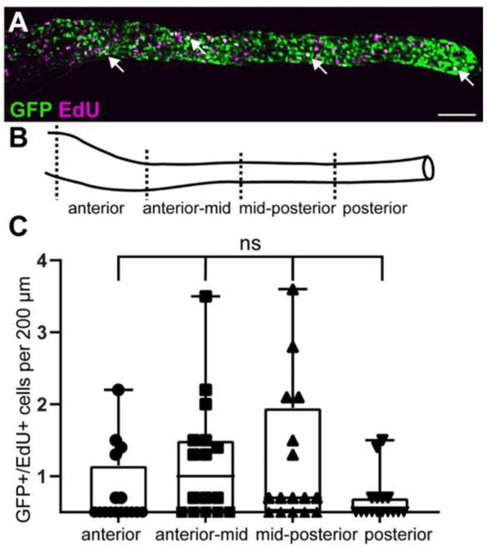
GFP+ cells include proliferating enteric progenitor cells at larval stages. (A) At 5 dpf, GFP colocalizes with EdU (white arrowheads). (B) Gut schematic indicates the four gut sub-regions analyzed. (C) Quantification of GFP and EdU colocalization in four gut regions shown in (B). Using one-way Anova, we did not find significant differences between the different gut regions. Box plot shows GFP+/EdU+ cells (2 experiments, 16 larvae). (A): maximum projection of dissected gut. Scale bar = 100 µm. ns not significant. |
|
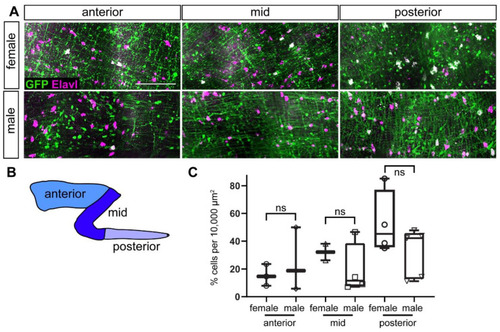
ENS neurons are GFP+ in adults. (A) GFP+ ENS neurons (white) at different anterior-posterior levels along the gut in male and female adult zebrafish. (B) Schematic of gut subdivisions. (C) Quantification of colocalization of GFP and Elavl in males and females in the three gut regions. Using t-tests, we did not find significant differences between males and females in the different gut regions. Box plot shows % GFP+/Elavl+ out of total Elavl cells per 10,000 µm2 (≥2 guts per condition). (A): maximum projections of dissected guts. ns not significant. Scale bar = 100 µm. |
|
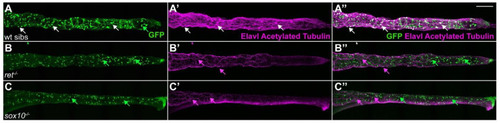
ret and sox10 mutants lack GFP+ ENS cells. (A–A”) In 6 dpf wildtype siblings (wt sibs), GFP+ cells colocalize with the pan-neuronal markers Elavl and acetylated Tubulin (AT, white arrows) in ENS neurons; Elavl−/AT−/GFP+ cells with a characteristic teardrop shape are enteroendocrine cells (green arrow). In 6 dpf ret (B–B”) or sox10 (C–C”) homozygous mutants, GFP+ ENS neurons are absent. AT+ processes are the vagal innervation to the gut (magenta arrows). Elavl−/AT−/GFP+ enteroendocrine cells are still present (green arrows). We analyzed ≥30 larvae per genotype. (A–C), maximum projections of dissected guts. Scale bar = 100 µm. |