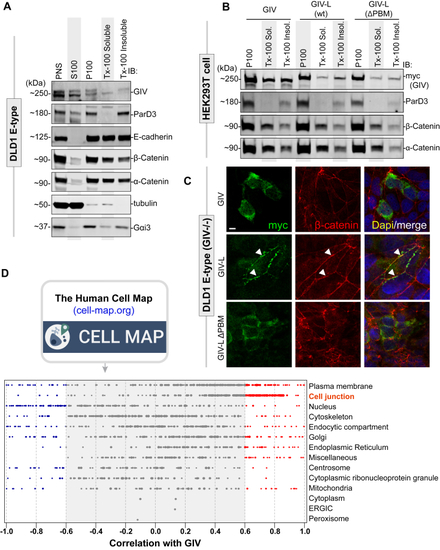
GIV-L, but not GIV, localizes at cell–cell junctions and its PBM is required for such localization.A, DLD1 E-type cells (parental or GIV-knockout) were fractionated into post nuclear supernatant (PNS), cytosolic (S100), crude membrane (P100), membrane detergent soluble (Tx-100 Soluble), and membrane detergent insoluble (Tx-100 Insoluble) pools. Equal proportions of each fraction were assessed for GIV by immunoblotting (IB). Equal loading and reasonable purity (lack of significant cross-contamination) of fractions were confirmed by immunoblotting for ParD3, E-cadherin, β-Catenin, α-Catenin, tubulin, and Gαi3. B, cell fractionation studies were carried out as in (A) on HEK293T cells exogenously expressing myc-tagged GIV, GIV-L (wt), or mutant GIV-L (ΔPBM). Equal proportion of each fraction was assessed for GIV and other loading and/or fractionation controls (as above) by immunoblotting (IB). C, DLD1 E-type cells were transfected with myc-tagged GIV, GIV-L (wt), or mutant GIV-L (ΔPBM), methanol fixed, and stained with anti-myc (green) or anti-β-Catenin antibody. Arrowheads indicate cell–cell contact sites. Scale bar, 7.5 μm. D, the publicly available BioID-based proximity map of HEK293T cells annotated in the Human Cell Map (HCM) was queried for GIV (without distinguishing GIV and GIV-L) and other preys that cotraffic and localize and functionally associate with GIV (i.e., prey–prey correlations) for various organelle-specific baits. The interactome is enriched for cell-junction-localized proteins.
|