- Title
-
Conserved And Non-Conserved Enhancers Direct Tissue Specific Transcription In Ancient Germ Layer Specific Developmental Control Genes
- Authors
- Chatterjee, S., Bourque, G., and Lufkin, T.
- Source
- Full text @ BMC Dev. Biol.
|
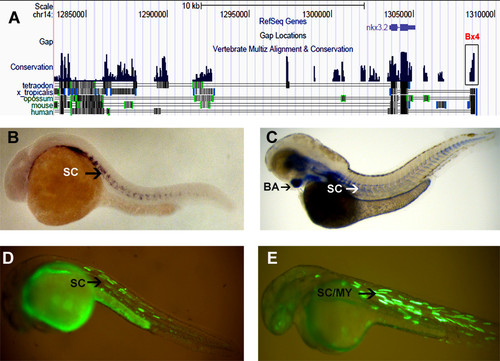
nkx3.2 CNEs and modified BAC. (A) UCSC browser snapshot showing the synteny around nkx3.2 and the functional CNE Bx4. (B) Whole mount RNA in situ hybridization showing expression of nkx3.2 in the sclerotome (SC) at 24 hpf. (C) Whole mount RNA in situ hybridization showing expression of nkx3.2 showing expression in branchial arch (BA) and sclerotome (SC) at 48 hpf. (D) Functional CNE Bx4 drives expression of EGFP in sclerotome (SC) at 24 hpf. (E) Modified BAC shows expression in both sclerotome and myotome (MY) at 24 hpf. |
|
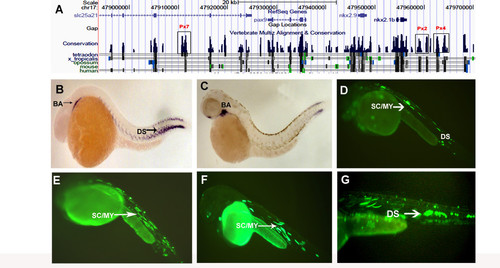
pax9 CNEs and modified BAC. (A) UCSC browser snapshot showing synteny around pax9 and the functional CNEs Px2, Px4 and Px7. (B) Whole mount RNA in situ hybridization showing expression of pax9 in branchial arch and dorsal sclerotome at 36 hpf and (C) 48 hpf. (D) Functional CNE Px2 drives expression of EGFP in sclerotome/myotome (SC/MY) as well as dorsal sclerotome at 36 hpf. Enhancers Px4 (E) and Px7 (F) drive expression in the sclerotome and myotome at 36 hpf and 48 hpf respectively. (G) Modified BAC showing expression in the dorsal sclerotome at 36 hpf. BA - branchial arch. DS - dorsal sclerotome |
|
pax9 non-conserved enhancer. (A) UCSC browser snapshot showing synteny around pax9 and the location of the functional CNE Px7 and the non conserved enhancer for branchial arch. (B) Whole mount RNA in situ hybridization showing expression of pax9 in branchial arch (BA) at 36 hpf. (C) A 500 bp non-conserved sequence drives expression of EGFP in the branchial arch at 36 hpf. EXPRESSION / LABELING:
|
|
otx1b CNES and modified BAC. (A) UCSC browser snapshot showing synteny around otx1b and the functional CNEs Ox2, Ox3 and Ox4. (B) Whole mount RNA in situ hybridization showing expression of otx1b in the neural plate at 6 hpf. (C) Whole mount RNA in situ hybridization showing expression of otx1b in the forebrain and otic vesicle at 24 hpf. (D) Modified BAC showing expression in forebrain and otic vesicle at 24 hpf. (E) CNE Ox4 drives expression in the neural plate at 6 hpf. CNE Ox2 (F) and Ox3 (G) drives expression in the forebrain at 36 hpf and 48 hpf respectively. Both fail to drive expression in the otic vesicle. FB-forebrain. OV-otic vesicle |
|
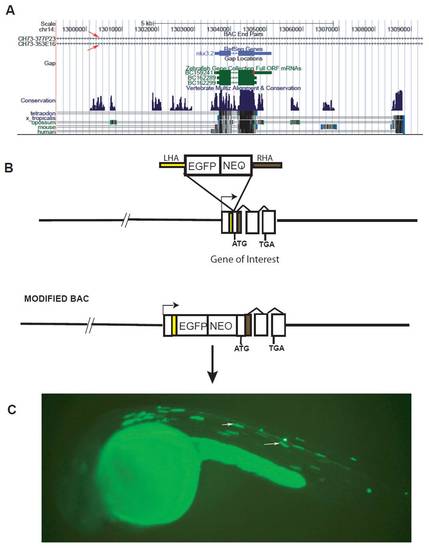
BAC modification process. (A) The UCSC genome browser showing two BACs (red arrows) spanning the gene nkx3.2 in zebrafish. (B) BAC modification by homologous recombination to insert a reporter gene and a drug selection cassette next to the translation start site of the gene (ATG). (C) A zebrafish carrying the modified BAC for the gene nkx3.2 expressing EGFP in sclerotomes (white arrows) at 24 hpf. Abbreviations: EGFP, enhanced green fluorescent protein; NEO, neomycin; LHA, left homology arm; RHA, right homology arm; TGA, stop codon. |
|
foxa2 CNEs and modified BAC. (A) UCSC browser snapshot showing synteny around foxa2 and the functional CNEs Fx2 and Fx6. (B) Whole mount RNA in situ hybridization at 48 hpf shows foxa2 expression in the rostral part covering forebrain and pharyngeal endoderm (C) Section in situ hybridization showing expression of foxa2 in forebrain at 48 hpf. (D) Section in situ hybridization showing expression of foxa2 in pharyngeal endoderm at 48 hpf. (E) Modified BAC showing expression in forebrain and (F) pharyngeal endoderm at 48 hpf.(G) CNE Fx2 and (H) Fx6 drives expression in the forebrain at 48 hpf. FB - forebrain. PE - pharyngeal endoderm |
|
BAC dissection to detect otx1b non-conserved enhancer. (A) The Otx1b 111 kb BAC showing the gene and the 6.4 kb non-conserved functional element and the CNEs present. The blue lines indicate the XbaI restriction sites. The plus sign in parenthesis indicates functional CNEs. (B) Six overlapping PCR fragments covering the 6.4 kb fragment. (C) Five overlapping PCR fragments covering the 1050 NC2 functional fragment (D) the seven Cy5 labeled probes designed on the functional NC2C fragment. Transgenic assay with the whole BAC (E), 6.4 kb fragment (F), NC2 (1050 bp) fragment (G) and NC2C (210 bp) fragment (H) all at 24 hpf. |
|
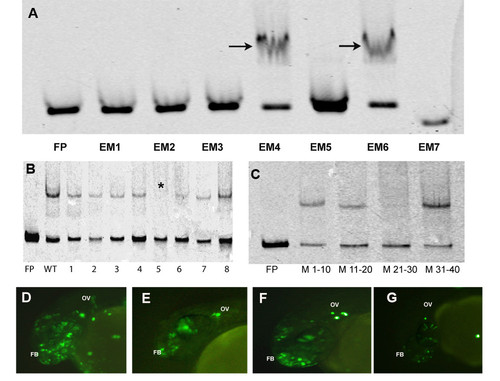
EMSAs and transgenic assays to detect core-binding motif. (A) Electrophoretic Mobility Shift Assays (EMSA) with 7 overlapping probes spanning the 200 bp non-conserved enhancer. Arrows indicate the two probes that bound protein. (B) EMSAs with 5 bp sliding mutant probes for EM4. The star indicates the probe 5 which did not show binding and the subsequent probe 6 showed weak binding. (C) EMSA with 10 bp mutation probes. Mutations in nucleotides 21-30 (M21-30) leads to complete abrogation of binding. Transgenic assays with wild type 40 bp probe (EM4) (D), mutant probe 5 (E), mutant probe 6 (F) and 10 bp mutant probe (G). The enhancer could drive expression in forebrain (FB) and otic vesicle (OV) at 24 hpf that are domains of expression of otx1b. |
|
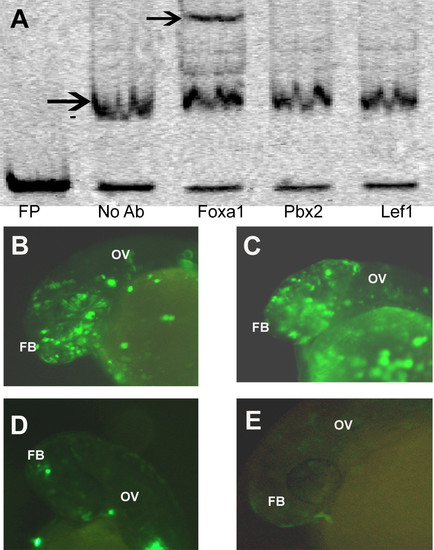
Elucidation of transcription factor binding to the enhancer. (A) Supershift assay with Foxa1, Pbx2 and Lef1 antibodies. Black arrow in no Ab lane denotes the shift and the one in Foxa1 added lane denotes supershift. (B) Expression of the 40 bp sequence (EM4) driven EGFP construct in forebrain (FB) and otic vesicle (OV) which are domains of expression of otx1b at 24 hpf. (C) Co-injection of the 40 bp enhancer along with a scrambled morpholino, showing no down regulation of the enhancer activity at 24 hpf. (D) Co-injection of the 40 bp enhancer along with morpholino 1 (M1) against Foxa1 showing reduction in the enhancer activity at 24 hpf. (E) Co-injection of the 40 bp enhancer along with morpholino 2 (M2) against Foxa1 showing reduction in the enhancer activity at 24 hpf. |
|
Immunohistochemistry for EGFP driven by nkx3.2 enhancer. (A) Enhancer Bx4 driven EGFP expresses in the sclerotomal cells (black arrow) at 24 hpf. As marked by nkx3.2 in the section RNA in situ hybridization (B) |
|
Immunohistochemistry for EGFP driven by pax9 enhancers. (A) Enhancer Px2 driven EGFP expresses in the sclerotomal cells (black arrow) at 36 hpf. The red arrow indicates background staining. Similar expression domains can be seen for enhancer Px4 (B) at 36 hpf and Px7 (C) at 48 hpf. (D) the negative control (empty vector injection) shows no staining in the sclerotome. (E) Section RNA in situ hybridization for pax9 at 36 hpf. |