- Title
-
EphA4 and EfnB2a maintain rhombomere coherence by independently regulating intercalation of progenitor cells in the zebrafish neural keel
- Authors
- Kemp, H.A., Cooke, J.E., and Moens, C.B.
- Source
- Full text @ Dev. Biol.
|
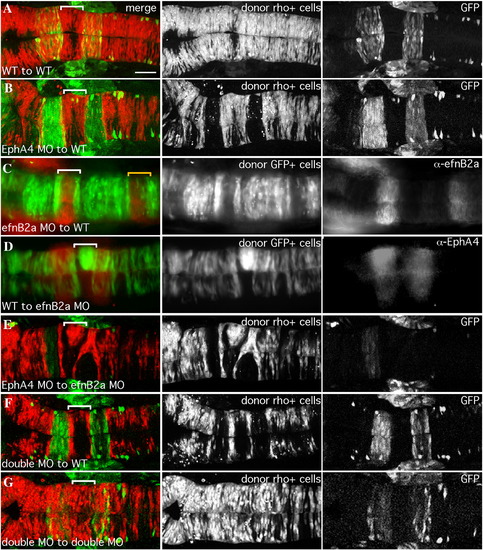
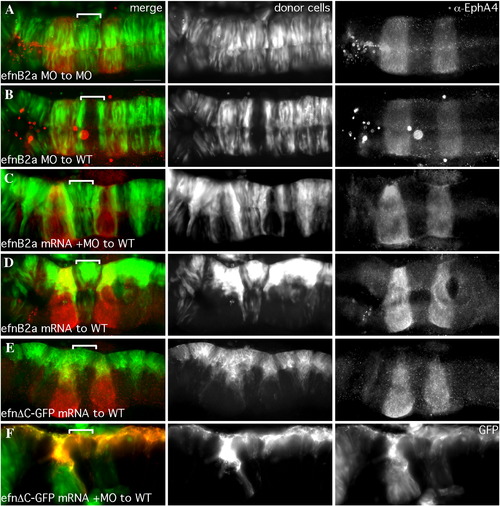
EfnB2a promotes cell adhesion in r4 independent of EphA4. 18 hpf mosaic embryos shown in dorsal view with anterior to the left. Left panels are merged images of individual channels shown in the middle and right panels. Donor cells (middle panel), are labeled with rhodamine dye (red in the merge; A, B, E, F, G) or express GFP (green in the merge; C, D). Rhombomere-specific markers (right panel): r3 and r5 are identified by expression of GFP in pGFP5.3 transgenic hosts (green in the merge; A, B, E, F, G) or detected by α-EphA4 (red in the merge; D); α-EfnB2a marks r1, r4, r7 (red in the merge; C). r4 is indicated by a white bracket in the merge. (A) WT cells contribute evenly to a WT host hindbrain. (B) EphA4 MO donor cells are excluded from r3 and r5 of a WT host. (C) EfnB2a MO donor cells are excluded from r4 and r7 (yellow bracket) of a WT host embryo. (D) WT donor cells form unilateral clusters on one side of r4 of an EfnB2a MO host. (E) EphA4 MO donor cells are excluded from r3 and r5 and form a unilateral cluster in r4 of an EfnB2a MO host embryo. (F) EfnB2a; EphA4 double MO donor cells are excluded from r3, r4, and r5 of WT host embryos. (G) EphA4; EfnB2a double MO donor cells contribute homogeneously to the hindbrain of a double MO host embryo. Scale bar: 50 μm. |
|
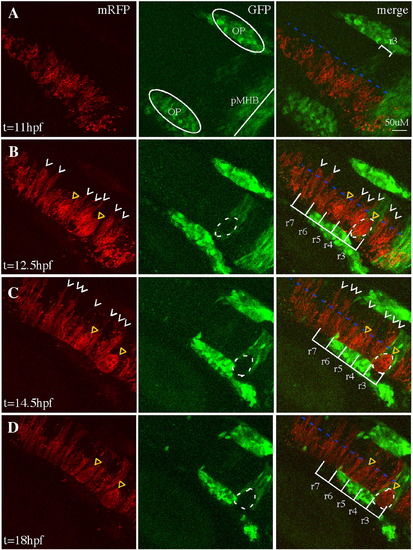
Cell sorting in EphA4MO mosaics begins during the neural keel stage. Representative 30 μm-deep projections of adjacent confocal sections through the hindbrain of a live mosaic embryo are shown at the indicated time points. Left panel: distribution of mRFP+ WT donor cells. Middle panel: expression of GFP in r3, r5 of the EphA4 MO pGFP5.3 transgenic host. Right panel is a merge of two channels shown at left. (A) At 11 hpf (neural plate), cells are uniformly distributed on one side of the hindbrain. (B) At 12.5 hpf (neural keel), presumptive daughters of neural progenitor divisions (white arrowheads) have begun to cross the midline (blue dotted line) but none is observed crossing in the r3 or r5 interior. (C, D) By 14.5 hpf (neural rod) and 18 hpf (neural tube), many donor cells have successfully crossed the midline (white arrowheads) except in r3 and r5, where WT mRFP+, GFP- cells form unilateral cell clusters (yellow triangles). OP (otic placode); pMHB (presumptive midbrain–hindbrain boundary). Scale bar: 50 μm. |
|
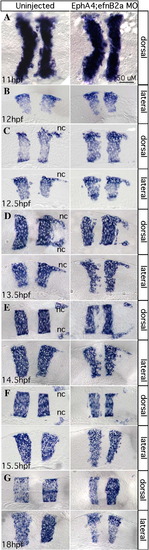
EphA4 and EfnB2a mediate hindbrain boundary sharpening during the neural keel stage. Whole mount (A) and 10 μm plastic sections (B–G) of RNA in situ hybridizations with a krox20/egr2b riboprobe. Anterior is to the left, dorsal or lateral views are indicated. Left panels show uninjected control embryos; right panels show stage-matched EphA4; EfnB2a double MO embryos. (A) Neural plate stage (N = 20); (B) neural plate to neural keel transition; (C,D) successive neural keel stages; (E,F) successive neural rod stages; (G) neural tube stage. Nc: r5-derived neural crest. Representative sections for each timepoint were chosen from at least 8 identically treated embryos. Scale bar: 50 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
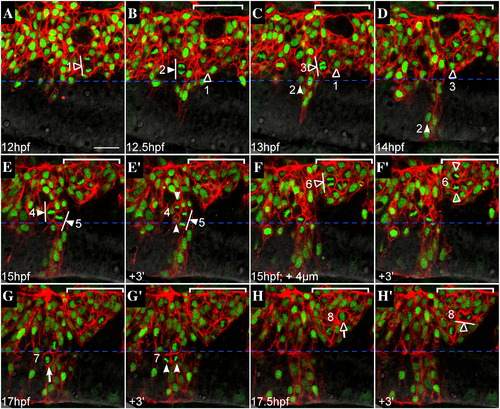
Failure to generate bilateral clones is not due to a defect in oriented cell divisions. Representative confocal sections from a time-lapse of a mosaic embryo, dorsal views, anterior to the left. WT donor cells (expressing mRFP to visualize cell membranes, histone-GFP to visualize nuclei) were transplanted into an EphA4MO pGFP5.3 host embryo (expressing GFP in r3 and r5; not shown). Midline is indicated by horizontal dashed blue line. r5 is indicated by white bracket in upper right hand corner of most images. Cells entering mitosis (rounded up; condensed chromatin) are indicated by white (r4) or hollow (r5) arrows. Cells in mitosis are indicated by arrowheads ending in perpendicular lines that represent spindle orientation. Daughters of numbered divisions that are still in the plane of focus are indicated by arrowheads. (A) At the onset of neural keel stage (12 hpf), donor cells are evenly distributed up to the midline throughout the hindbrain. Because the transgene is not expressed in r5 at this early timepoint, r4 and r5 territories cannot be definitively assigned. However a cell that is unambiguously assigned in later timepoints to r5 has already entered anaphase and is positioned close to the midline (hollow arrowhead; division “1”). (B) At early neural keel (12.5 hpf), a WT donor cell in r4 divides with a medial nuclear position and transverse spindle orientation (white arrowhead; division “2”). The more medial daughter from division “1” is located in r5. (C) At 13 hpf a donor cell in r5 divides medially with transverse spindle orientation (hollow arrowhead; division “3”). Meanwhile, the more medial daughter of division “2” has begun to cross the midline (white arrowhead). The other daughter from division “2” remains on the transplanted side of the neuroepithelium throughout the timelapse (different plane of focus; data not shown). (D) By 14 hpf, one daughter from division “2” (white arrowhead) has completed midline crossing and is integrated into the contralateral side of the neuroepithelium, whereas the more medial daughter from division “3” (hollow arrowhead) remains on the original side of the neuroepithelium. Both daughters of division “3” were tracked for an additional 2 h and failed to cross the midline during this time (data not shown). (E) At 15 hpf, two WT donor cells in r4 with medially positioned nuclei are in metaphase with mitotic spindles orientated transverse to the midline (white arrowheads; divisions “4” and “5”). (E′) Within 3 min, a dividing cell from panel C has entered telophase (white arrowhead; division “5”) and another has completed cytokinesis, generating a more medial and more lateral daughter cell (white arrowheads; division “4”). (F) At the same timepoint as in panel E (15 hpf), but 4 μm deeper into the embryo, a laterally displaced WT donor cell in r5 (hollow arrowhead; division “6”) is shown at anaphase with a transverse spindle orientation (white line). (F′) Within 3 min, the dividing cell from panel F has completed cytokinesis, generating a more lateral and a more medial daughter cell (hollow arrowheads), neither of which crosses the midline during the course of the timelapse (data not shown). (G) WT donor cell in r4 at the neural tube stage (17 hpf) rounds up prior to division with medial nuclear position (white arrow; division “7”). (G′) Donor cell indicated in panel G has completed cytokinesis. Daughters of this division are born side by side, equidistant from the midline (white arrowheads). (H) Laterally displaced WT donor cell in r5 rounds up and has condensed chromatin prior to mitosis (hollow arrow; division “8”). (H′) Within 3 min, the dividing donor cell from panel H has entered telophase. Spindle orientation is planar, ie. parallel to the midline (white line). Scale bar: 25 μm. |
|
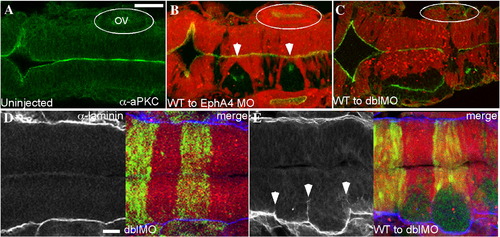
Failure to execute cross-midline divisions results in duplicated neuraxes in EphA4 and EfnB2a MO mosaics. Immunostaining of 18 hpf mosaic embryos shown in dorsal view with anterior to the left. (A–C) merged images showing α-aPKC (green) and MO host cells labelled with rhodamine (red). (A) aPKC is expressed on the apical surface of neuroepithelial cells, which is at the midline. (B) WT EphA4+ donor cells (unlabelled) fail to cross the midline and form unilateral clusters in r3 and r5 when transplanted into an EphA4MO host (white arrowheads); these clusters have an ectopic aPKC+ apical region. Likewise WT EfnB2a+ donor cells form a unilateral cluster in r4 of an EfnB2a MO host (data not shown). (C) WT cells transplanted into an EphA4; EfnB2a double MO host form three separate clumps with ectopic apical surfaces (data not shown) or fuse to give rise to a partially duplicated neuroepithelium in the r3–r5 region; cell clustering is also sometimes evident in r7 (N=6). OV (otic vesicle). (D,E) α-laminin staining (left panels); right panel is a merge showing the rhodamine-labeled double MO host cells (red); host cells contain the pGFP5.3 transgene and express GFP in r3 and r5 (green). (D) Laminin is expressed along the lateral edges of the neuroepithelium in WT (not shown) and in double MO host embryos (N=24). (E) Laminin is ectopically expressed around the edges of the cluster of unlabelled WT cells in a WT to double MO mosaic embryo (N=27). Scale bars: 50 μm. |
|
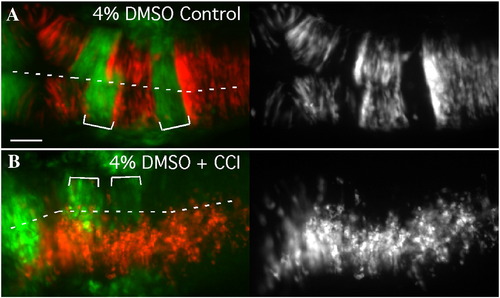
Cell sorting is prevented by inhibiting cell division in the neural keel/rod. Dorsal views of 18 hpf mosaic embryos with anterior to the left. (A) EphA4 MO donor cells (red) sort out of r3 and r5 of a WT pGFP5.3 host (green) treated with solvent alone (4% DMSO; N = 10). (B) Treatment of mosaic embryos from 90% epiboly onwards with cell cycle inhibiting drugs inhibits cross-midline divisions and also prevents sorting-out of EphA4MO cells from r3 and r5 (N = 7/11). Scale bar: 50 μm. |
|
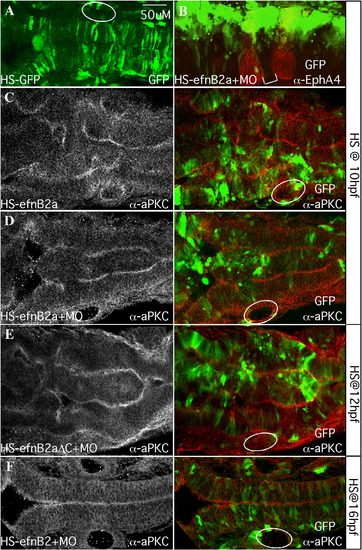
Mosaic EfnB2a overexpression in the neural keel causes widespread cell sorting and duplication of the neuraxis. Dorsal views of 18–20 hpf embryos injected with heat-shock (HS)-GFP (A) or HS-EfnB2a DNA constructs (B–F) as indicated. Anterior is to the left. White circles indicate the position of the otic vesicle. (A) pHS-GFP-injected embryo coinjected with EfnB2a MO, and heat shocked at 10 hpf. GFP+ cells are symmetrically distributed on both sides of the neuroepithelium (N = 12/12). (B) pHS-EfnB2a-HS-GFP injected embryo coinjected with EfnB2a MO, heat shocked at 10 hpf and stained with α-EphA4 to detect r3, r5 (red). r4 is indicated with a white bracket. EfnB2a+/GFP+ cells (green) are distributed asymmetrically throughout the hindbrain, including in r4 (N = 16/27). (C–F) Embryos injected with the indicated HS constructs, with or without EfnB2a MO, heat shocked at the timepoint indicated on the far right and stained with α-aPKC. Left panel: aPKC expression alone; right panel: merged image showing distribution of EfnB2a+/GFP+ cells (green) and aPKC expression (red). (C, D) Embryo injected with pHS-EfnB2a-HS-GFP in the absence or presence of coinjected EfnB2a MO, heat shocked at 10 hpf. aPKC expression in two parallel stripes reveals partial duplication of the neuraxis (N = 7/14, and 9/14, respectively). (E) Embryo injected with the HS-EfnB2a-ΔC allele and EfnB2a MO, heat shocked at 12 hpf also shows neuraxis duplication (N = 9/20). A similar phenotype is observed when embryos are injected with the WT EfnB2a allele HS construct, in the absence or presence of EfnB2a MO, and heat shocked at 12 hpf (data not shown; N = 3/7, and 5/8, respectively). (F) Embryo co-injected with pHS-EFnB2a-HS-GFP and EfnB2a MO, then heat shocked at 16 hpf. EfnB2a-expressing cells contribute homogeneously to the epithelium and no duplication of the neuroepithelium has occurred (N = 14/14). Immuno-staining confirms that EfnB2a expression is still ectopically induced in embryos heat shocked at 16 hpf (data not shown; N = 7/7). Scale bar: 50 μm. |
|
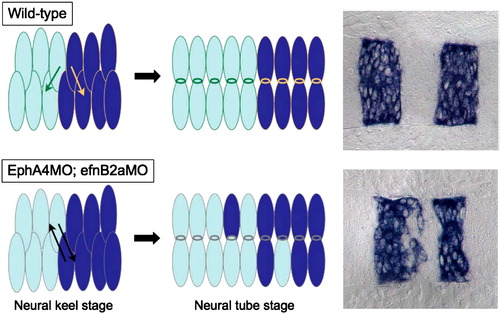
A model: unrestricted cross-midline divisions disrupt rhombomere boundaries in an EphA4; EfnB2a MO embryo. In a WT hindbrain, daughter cells created by cross-midline divisions in r3, r4 and r5 are restricted by the presence of cell surface EphA4 and EfnB2a to intercalate well inside their respective rhombomeres of origin. Neuroepithelial cells in r3–5 in embryos lacking EphA4 and EfnB2a divide without such restrictions, leading to mis-sorting of daughters from r3 and r5 mothers into r4 and vice versa. |
|
efnB2a mRNA expression rescues cell–cell adhesion in EfnB2a MO r4, while over-expression induces adhesion-based clustering. Dorsal views of 18 hpf embryos with anterior to the left. r4 is indicated by a white bracket. Left hand panels are merged images of individual channels. Middle panel: Donor cells, pseudocolored in green (A–E) or red (F) in the merge, express GFP (A–E) or memb-RFP (F) from injected mRNA. r3 and r5 are visualized by α-EphA4 staining (red), or by the expression of GFP in r3 and r5 of a transgenic pGFP5.3 host. (A) Cells depleted of Efnb2a by MO injection contribute normally to an EfnB2a MO host embryo. (B) Cells depleted for EfnB2a by MO are excluded from r4 of a WT host. (C) EfnB2a MO donor cells coinjected with EfnB2a mRNA contribute to r4 of WT hosts. Ectopically expressing EfnB2a+ donor cells are simultaneously excluded from rhombomeres that do not normally express EfnB2a and instead express Eph receptors (r2,3,5, and r6). (D) WT donor cells injected with a “rescuing” amount of EfnB2a mRNA form unilateral cell clusters in r4 of a WT host. (E) WT donor cells expressing the EfnB2aΔC-GFP allele, in which the C-terminus is replaced with GFP, fail to cross the midline in r4 of a WT host. Note that this GFP fusion protein is localized to cell membranes, as expected. (F) Efnb2a MO donor cells expressing the EfnB2aΔC-GFP allele also fail to cross the midline in r4 of a WT host. Note that donor cells are visible in the GFP channel, confirming expression of the truncated allele. Scale bar: 50 μm. |
|
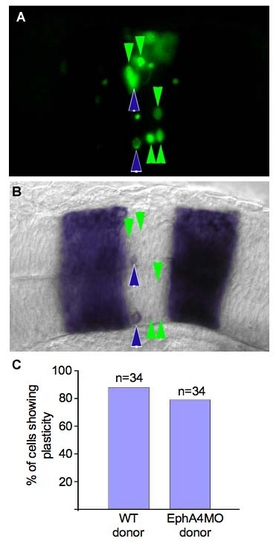
Mis-localized hindbrain cells exhibit plasticity independently of EphA4. (A,B) dorsal views of the same 18 hpf embryo. (A) Green cells were transplanted from r3 of a donor embryo into r4 of a host embryo at the 10–12 somite stage. (B) RNA in situ hybridization showing krox20/egr2b expression. Green arrows indicate cells that have changed their identity, blue arrows indicate cells that have retained r3 identity. (C) EphA4 is not required for plasticity. The proportion of cells that turn off krox20/egr2b under these transplantation conditions is unaffected if the transplanted cells lack EphA4. |
|
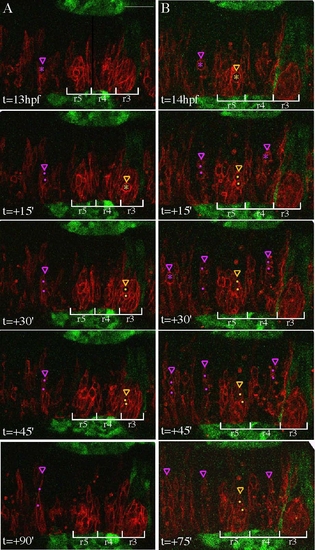
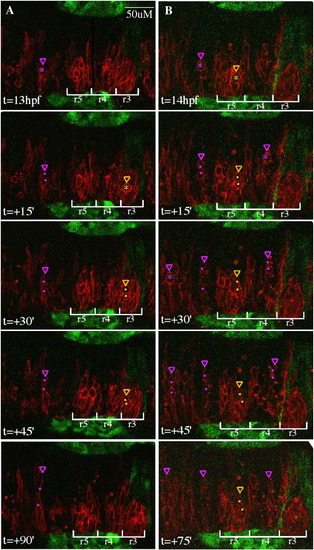
Confocal time-lapse of memb-RFP+ WT cells transplanted into an EphA4MO pGFP5.3 host embryos (GFP in r3, r5, midbrain/hindbrain boundary, and otic placodes/vesicles), showing that donor cells divide with similar timing irrespective of rhombomere location. Dorsal views with anterior to the right. Transient cell rounding, a precursor to cell division, is observed in neural progenitors throughout the hindbrain at neural keel stages. (A,B) Representative confocal sections of the zebrafish hindbrain are shown starting at 13 hpf or 14 hpf, respectively, and every 15–39′ thereafter. Cells about to undergo division in r3 or r5 are marked by yellow asterisks, their daughters are marked with yellow dots, and the most medial extension of the dividing mother or the more medial daughter is indicated by a yellow arrowhead. Cells are similarly annotated in purple in other rhombomeres. During the 75–99′ time period shown, several cell divisions were completed in r3 and r5. Daughter cells from these divisions were not observed crossing the midline, although 1–2 h after division, many daughter cells either disappeared from the plane of focus or could not be unambiguously distinguished and therefore we were unable to determine their final locations. At the same time, neural progenitors divided in other rhombomeres and frequently the more medial daughter extended a projection toward the other side of the hindbrain, most often reaching at least halfway across to the other side within 99′ after division. Scale bar: 50 μm. |
|
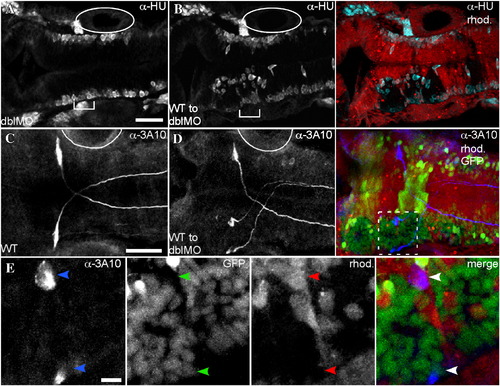
WT cell clusters are neurogenic. Immunostaining of embryos shown in dorsal view with anterior to the left using α-HU at 26 hpf (A,B), or α-3A10 at 30 hpf (C–E). (A) In an EphA4;EfnB2a double MO embryo, neurogenesis, indicated by α-HU immunostaining, takes place at the lateral surfaces of the neuroepithelium. (B) Left panel shows α-HU immunostaining. Right panel is a merge showing rhodamine-injected double MO host cells surrounding unlabeled WT cell clusters. Ectopic neurogenesis is apparent at the edges of WT cell clusters (cyan). (C) α-3A10 shows the bilateral Mauthner neuron pair in r4 of a 30 hpf WT embryo. (D) Left panel shows α-3A10 immunostaining. Right panel is a merge of 3A10 staining in blue with rhodamine-injected double MO host cells (red) that also contain the noiGFP transgene (yellow) surrounding clusters of histone-GFP+ nuclei from WT donor cells (green). In WT to double MO mosaic embryos, ∼ 15% of embryos show duplication of the Mauthner neuron (N = 3/19). (E) Closeup view of the boxed region in D shows that one of the ectopic Mauthner neurons in r4 is donor-derived (histone-GFP+ nucleus, rhod.-negative cytoplasm). Scale bars: 50 μm (A–D), or 10 μm (E). Otic vesicle, adjacent to r5, is indicated by full or partial oval. |
|
|
Reprinted from Developmental Biology, 327(2), Kemp, H.A., Cooke, J.E., and Moens, C.B., EphA4 and EfnB2a maintain rhombomere coherence by independently regulating intercalation of progenitor cells in the zebrafish neural keel, 313-326, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.