- Title
-
Chemical Genetics Screen Identifies Epigenetic Mechanisms Involved in Dopaminergic and Noradrenergic Neurogenesis in Zebrafish
- Authors
- Westphal, M., Sant, P., Hauser, A.T., Jung, M., Driever, W.
- Source
- Full text @ Front Genet
|
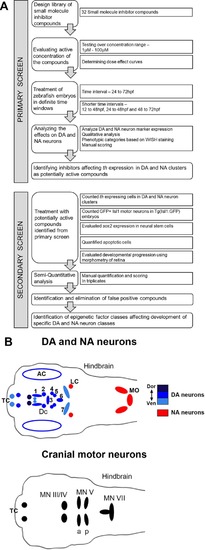
Design of Small Molecule Screen in two phases. |
|
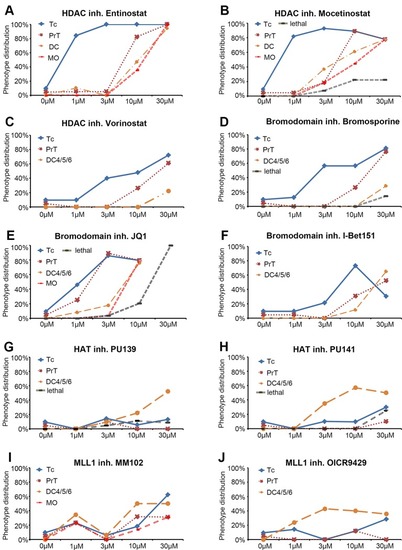
Dose response curves for selected small molecule inhibitors. |
|
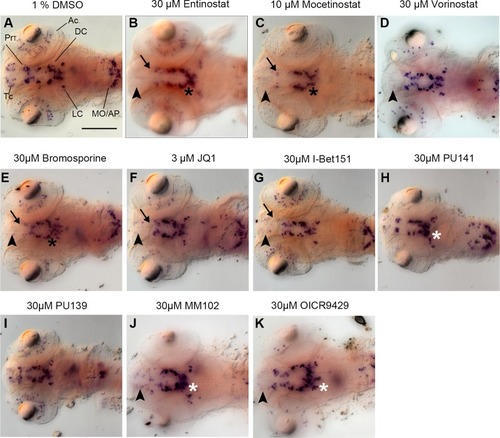
Primary Screen results for representative small molecule inhibitors. |
|
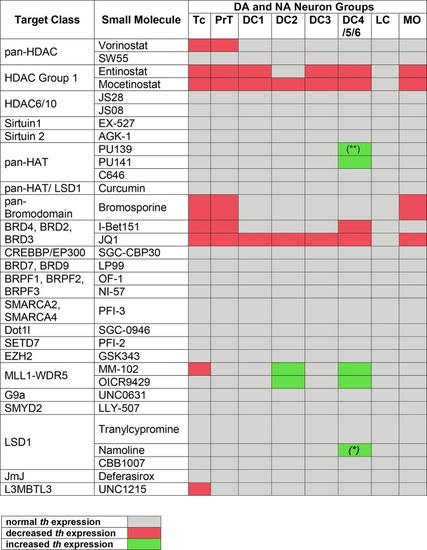
Summary of Primary Screening Results. Table depicting effects on |
|
Quantification of Secondary Screen—HDAC inhibitors. |
|
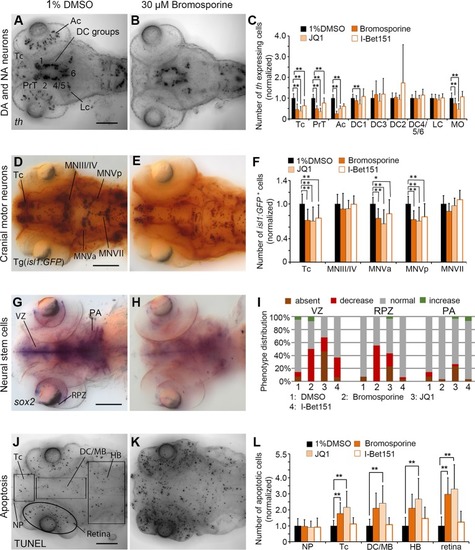
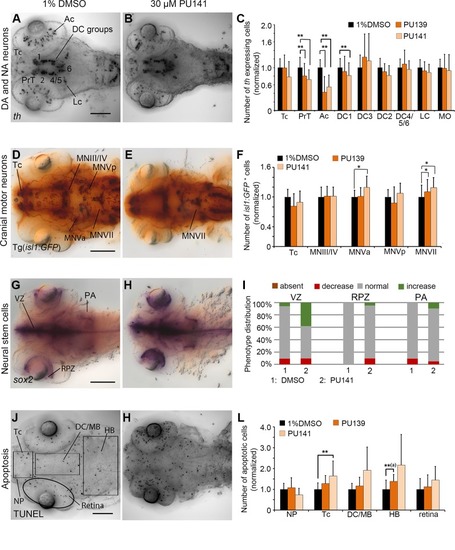
Quantification of Secondary Screen—Bromodomain inhibitors. |
|
Quantification of Secondary Screen, HAT inhibitors. |