Fig. 1
- ID
- ZDB-FIG-240510-35
- Publication
- Ou et al., 2024 - Formation of different polyploids through disrupting meiotic crossover frequencies based on cntd1 knockout in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
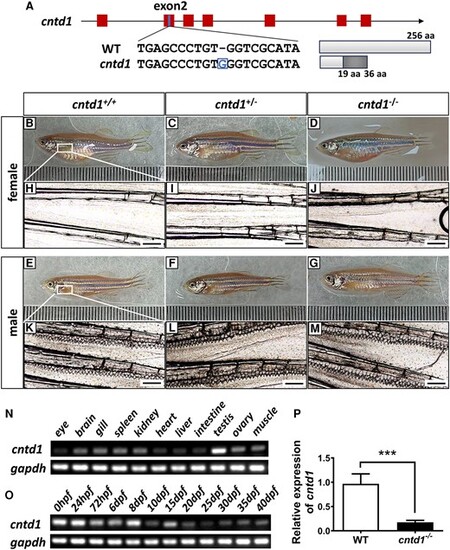
Generation of cntd1 knockout zebrafish. A, Schematic representation of the genomic structures of the zebrafish cntd1 gene and the corresponding putative peptides and the target sites of engineered CRISPR-Cas9 in exon 2 (exons shown in red). The inserted base is highlighted in the blue box. The correct amino acids are indicated by white boxes, while the incorrect amino acids in cntd1 mutants are presented in a dark box. B–G, General morphological observations of zebrafish with different genotypes: cntd1+/+ (B), cntd1+/− (C), and cntd1−/− (D) females; cntd1+/+ (E), cntd1+/− (F), and cntd1−/− (G) males. H–M, Microscopic observation of the central fin rays on the pectoral fins. Absence of breeding tubercles in cntd1+/+ (H), cntd1+/− (I), and cntd1−/− (J) females; the presence of spike-like breeding tubercle clusters in cntd1+/+ (K), cntd1+/− (L), and cntd1−/− (M) males. Scale bar: 200 μm. (N) Tissue distribution of endogenous cntd1 transcripts with RT-PCR in WT zebrafish. (O) Expression levels of endogenous cntd1 at different developmental stages checked by RT-PCR in WT zebrafish. (P) Relative expressions of cntd1 in the testis of WT and cntd1−/− zebrafish. Six individuals were used, and ef1α was selected as the internal reference in this experiment. The error bars represent the means ± SDs; “***” above the error bar indicates statistically significant differences at P < 0.001 by two-tailed Student's t-test. |