Fig. 3
- ID
- ZDB-FIG-240109-98
- Publication
- Song et al., 2023 - Structural basis for inactivation of PRC2 by G-quadruplex RNA
- Other Figures
- All Figure Page
- Back to All Figure Page
|
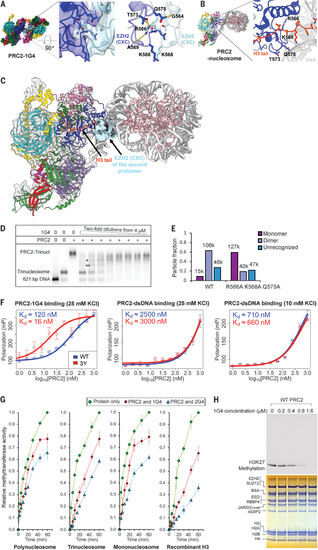
RNA-mediated PRC2 dimer is an inactive complex. (A) Cryo-EM structure of PRC2-1G4 complex with zoom-in to show the interface of two EZH2 CXC domains. Interacting residues (R566, K568, T573, and Q575) are highlighted in the stick representation. One set of hydrogen bonds (R566NE-T573OG1, 2.70-Å distance; R566NH1-A569O, 2.73-Å distance; and Q575NE2-G564O, 2.51-Å distance) is indicated by yellow dashed lines. The other set of hydrogen bonds between the same amino acid pairs in the second PRC2 protomer is not shown for clarity. Another view of the same region is shown in fig. S3G. (B) Structure of PRC2-nucleosome complex (PDB: 6WKR) with zoom-in to emphasize the CXC interactions with nucleosome H3 tail (orange). The same residues as shown in (A) are highlighted in stick representation. (C) Superposition of the EZH2 CXC domain of the RNA-bound PRC2 on the nucleosome-bound PRC2 to emphasize disparate functions of the CXC domain. (D) Nucleosome-RNA competition assay. PRC2 was incubated with constant amount of radiolabeled trinucleosome and serial dilutions of 1G4 RNA. Incomplete PRC2-trinucleosome complexes are indicated by * and **. We assume two of three nucleosomes were occupied by PRC2 in *, and one of three nucleosomes in **. (E) Negative-stain EM to quantify monomer and dimer particles of EZH2 R566A K568A Q575A binding 1G4. The number of particles in each class is indicated above each bar. (F) Binding affinity of 1G4 RNA or dsDNA to WT PRC2 and EZH2 R566Y K568Y W575Y (3Y) measured by FP. We used a reaction buffer with a lower salt concentration to achieve higher binding affinity for the dsDNA (right). Kd, dissociation constant. (G) Methyltransferase activities from figs. S12 to S15 are plotted against reaction times for PRC2 (400 nM) preincubated with 1G4 (400 nM), 2G4 (400 nM), and mock (protein only). Error bars represent standard deviations of three replicates performed on different days. (H) Methylation of trinucleosomes by PRC2 with serial dilutions of 1G4 RNA. (Top) Radiogram that shows methylation. (Bottom) Coomassie-stained gel. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. |