Fig. 1
- ID
- ZDB-FIG-240109-96
- Publication
- Song et al., 2023 - Structural basis for inactivation of PRC2 by G-quadruplex RNA
- Other Figures
- All Figure Page
- Back to All Figure Page
|
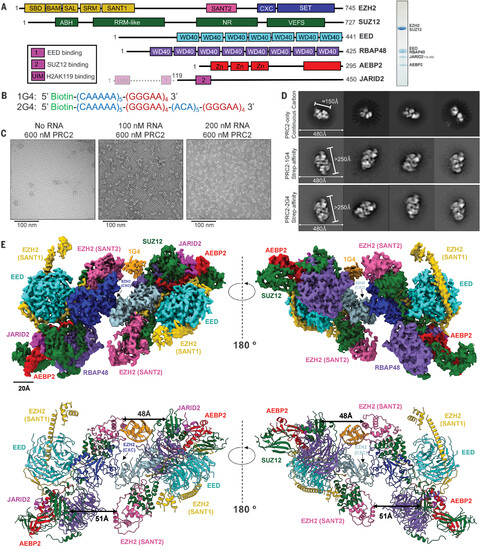
Overall structure of a PRC2-1G4 RNP complex. (A) (Left) Schematic of the proteins in the PRC2.2 six-subunit complex. Transparent N-terminal region of JARID2 was not included. (Right) Coomassie-stained gel of purified PRC2. (B) The two RNA oligonucleotides used in this study. (C) Negative-staining EM images of streptavidin-affinity grids with excess PRC2 and various 1G4 RNA concentrations. (D) Negative-staining EM provided 2D-class averages of PRC2 alone collected from continuous carbon grids and PRC2-G4 RNAs from streptavidin-affinity grids. (E) (Top) Cryo-EM density map of PRC2 bound to 1G4 RNA. EZH2 (CXC-SET) of protomer 1 in blue, EZH2 (CXC-SET) of protomer 2 in light blue, and 1G4 RNA in orange. (Bottom) The atomic model of PRC2 bound to 1G4 RNA. Distances between EZH2 (SANT2) and SUZ12 (RRM-like) are highlighted by black arrows to emphasize the change from 1G4-binding. |