FIGURE 6
- ID
- ZDB-FIG-220722-22
- Publication
- Mukaigasa et al., 2021 - The developmental hourglass model is applicable to the spinal cord based on single-cell transcriptomes and non-conserved cis-regulatory elements
- Other Figures
- All Figure Page
- Back to All Figure Page
|
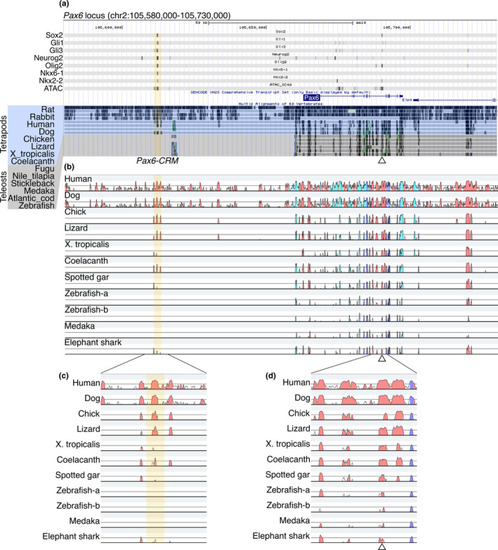
Identification of Pax6-CRM and its diversification in vertebrates. (a) ChIP-seq and ATAC-seq peak call results are displayed in the UCSC genome browser with Multiz Alignments track (the display mode is dense). The mouse Pax6 locus is displayed. Regions harboring multiple TF binding sites (Pax6-CRM) are highlighted in yellow, and a previously validated CRM is indicated by an open triangle under the track (Oosterveen et al.,2012). (b) The same genomic regions from several species are aligned, and sequence conservation is visualized by VISTA. The base sequence is mouse, and the species compared are indicated on the left side. The peaks of the conserved regions are colored pink (noncoding sequences), dark blue (exons), or light blue (UTRs). Zebrafish possess two pax6 genes (pax6a and pax6b); thus, both loci are included in the alignments (Zebrafish-a and Zebrafish-b correspond to the pax6a and pax6b loci, respectively). (c, d) Enlarged view of the plot focusing on CRMs. The previously identified CRM located in the intron is conserved in all species examined (d), whereas the Pax6-CRM highlighted in yellow is not conserved in zebrafish and medaka (c) |