|
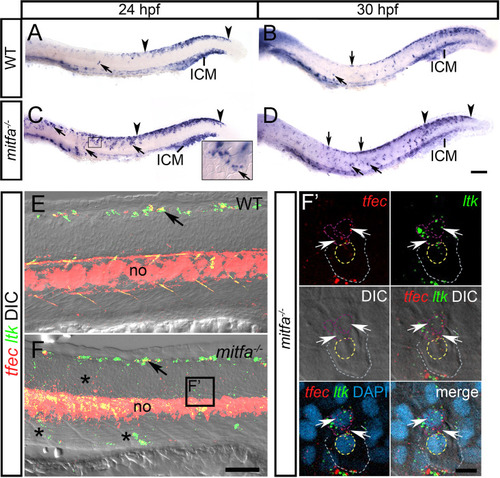
Mitfa represses <italic>tfec</italic> expression in melanoblasts.In WT embryos at 24 hpf (A) and at 30 hpf (B), tfec expression is detectable in Ib(sp) and Ib(df) (arrows), respectively, and in the posteriorly regressing early NCC/Cbl domain of the dorsal posterior trunk and tail (region within arrowheads), but expression is undetectable in the lateral migration pathway. At 24 hpf (C) and at 30 hpf (D), mitfa mutants present with an increased number of tfec-positive cells (arrows) along the dorsal trunk, as well as in the medial and lateral (C, inset) migratory pathways. RNAscope (E,F) performed on mitfa mutants at 30 hpf shows an increased number of tfec-positive cells compared to the WT on the migration pathways (F, asterisks). Arrows indicate iridoblast precursors along the dorsal trunk. (F’) RNAscope reveals that in mitfa mutants ectopic tfec-positive cells migrating on the lateral migration pathway (below the epidermis; grey and yellow dashed lines indicate the periphery and nuclear boundary, respectively, of overlying keratinocytes) co-express ltk (arrows; nuclei indicated by purple dashed lines), and thus likely correspond to Ib(sp). ICM, intermediate cell mass; LP, lateral patches; no, notochord. Lateral views, head towards the left. Scale bars: A-D: 100 μm; E,F: 50 μm; F’: 10 μm.
|