Fig. S5
- ID
- ZDB-FIG-120720-80
- Publication
- Zou et al., 2012 - Crb Apical Polarity Proteins Maintain Zebrafish Retinal Cone Mosaics via Intercellular Binding of Their Extracellular Domains
- Other Figures
- All Figure Page
- Back to All Figure Page
|
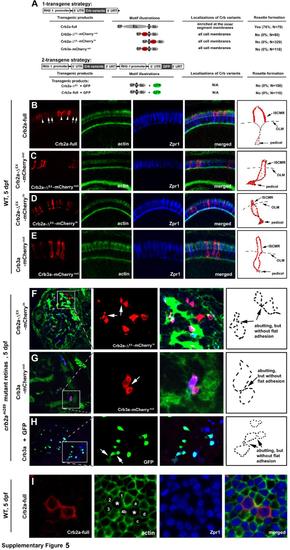
The extracellular domain of Crb2a is required for the localization of Crb2a to the inner segment cell membranes and for photoreceptor adhesion in zebrafish retinas, related to Figure 6. A. The schematics illustrate the two strategies to transgenically express Crb2a, Crb2a-ΔEX-mCherryout, Crb2a-ΔEX-mCherryin, Crb2a-ΔEX, Crb3a-mCherryout, or Crb3a in zebrafish retina. Individual protein domains are labeled as follows: SP, the signal peptide; Ex., the extracellular domain; T, the transmembrane domain; In., the intracellular domain; mC, mCherry; GFP, green florescent protein; -ΔEX, absence of the extracellular domain. The percentages of transgenic photoreceptors that formed rosettes in 5-dpf crb2am289 mutant retinas were quantified and summarized in the table. In addition, the subcellular localizations of the transgenic products in wildtype retina (vertical sections for B-E; transverse section for I) were summarized in the table as well. B-E. Wildtype full-length Crb2a (arrowheads) was highly enriched at the inner segment cell membranes in the photoreceptors (B), whereas Crb2a-ΔEX-mCherryout (C), Crb2a-ΔEX-mCherryin (D), and Crb3a-mCherryout (E) localized diffusively to the entire cell membranes of the transgenic photoreceptors. Due to the strong expression by the RH2-1 promoter, endogenous Crb2a (white arrows) appeared much weaker under exposure for better revelation of transgenically-expressed Crb variants. Schematic drawings in the right panels illustrate the membrane distribution patterns of Crb variants in a representative transgenic cone. F-H. Expression of the extracellular domain-lacking Crb variants Crb2a-ΔEX-mCherryin (F), Crb3a-ΔEX-mCherryout (G), or untagged Crb3a along with GFP (H) did not prompt photoreceptor aggregation in crb2am289 mutant embryos at 5 dpf. The leftmost panels show the entire retinal sections, in which the boxed local regions were magnified and presented in the middle panels for better illustration. Arrows indicate the abutting cells that did not adhere to each other to form a flat junctional interface as in Fig. 6H. Schematic drawings in the right panels illustrate the contours of representative transgenic cells that are abutting but not adhering to each other. Green staining is for actin, except as indicated. Double cones are shown in blue. Crb variants are in red. I. Overexpression of RH2-1 promoter-driven full-length wildtype Crb2a increased the sizes of the inner segments of transgenic cones. To quantify the increase in the sizes of the inner segments, the area of the transverse section of Zpr1 positive transgenic cones at the level of OLM was compared with the average size of their non-transgenic Zpr1 positive neighbor cones. For example, the size of the left transgenic cell was compared with the average size of its neighbor cells1-4, and the right cell with its neighboring cells a-d. A total of 28 transgenic cells were analyzed with MetaMorph 7.0 software, showing an average of a 1.79 fold increase in the cross section sizes at the OLM level. Full-length Crb2a (Crb2a-full) was visualized by the anti-Crb2a97-457 antibody. A weak imaging exposure was used to better reveal the overexpression of transgenic Crb2a but not the endogenous Crb2a. |
Reprinted from Developmental Cell, 22(6), Zou, J., Wang, X., and Wei, X., Crb Apical Polarity Proteins Maintain Zebrafish Retinal Cone Mosaics via Intercellular Binding of Their Extracellular Domains, 1261-1274, Copyright (2012) with permission from Elsevier. Full text @ Dev. Cell