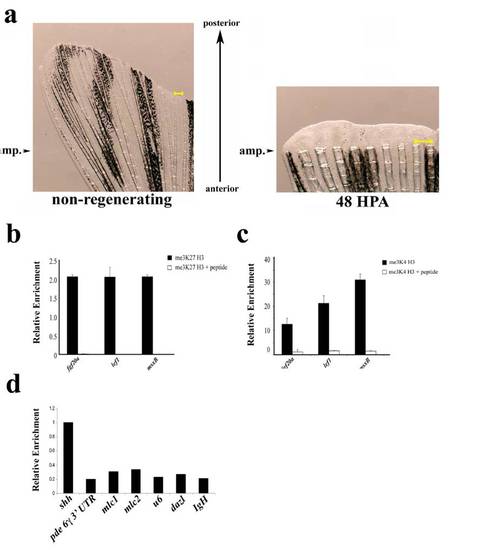
Fig. S2
|
(a) Examples of uncut and regenerating fins. Arrowheads indicate the site of amputation on an un-amputated fin (Left) or a fin at 48 h post amputation (Right). Tissue distal to these arrowheads was used for ChIP or gene expression analysis. The regenerating fin is magnified to allow visualization of the newly formed regenerating tissue and a scale bar (yellow) is shown in each panel. (b and c) Specificity of methyl histone ChIPs. me3K27 H3 ChIP (b) or me3K4 H3 ChIP (c) were performed from non-regenerating chromatin with the indicated antibodies that had been preincubated with either BSA (1 mg/mL) or peptide antigens (10χmolar excess). Preincubation of antibody with peptide significantly reduces amount of eluted material relative to control. (d) Examples of genomic regions of relatively high and low levels of me3K4 H3. me3K4 H3 ChIP was performed as above (without peptide competition) and eluates were examined by real-time PCR with the indicated primer sets. Data are expressed relative to the ChIP of the transcription start site of shah to highlight the significantly lower levels of me3K4 H3 at the other genomic positions. Primers used were: 3′ UTR of phosphodiesterase 6 gamma; cardiac myosin light chain 1 (mlc1) promoter; cardiac myosin light chain 2 (mlc2) promoter; u6 small RNA promoter; dazl promoter; IgH diversity region intronic sequence (IgH). |