Fig. S8
|
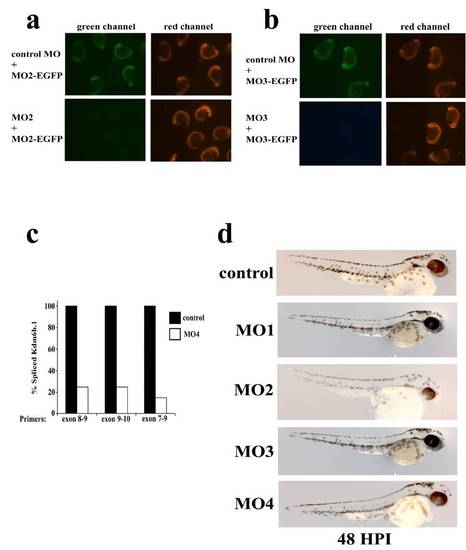
Morpholino control experiments. (a and b) To demonstrate efficacy of the morpholinos, the target sequence for MO2 and MO3 was cloned upstream of EGFP in pCS2 vector. Animals were injected with RNA transcribed from these vectors in the presence of mRFP RNA and the indicated morpholino. Twenty-four hpi animals were illuminated under epifluorescence and photographed. Exactly the same exposure times were used in each of the four panels shown. MO1 is a FITC labeled translation blocking morpholino which prevented us from performing the heterologous reporter assay as was done for MO2 and MO3. (c) In the case of MO4, real-time PCR was performed on cDNA derived from control or MO4 injected animals at 24 hpi. Primers were designed to amplify regions of the correctly spliced Kdm6b.1 mRNA around exon 8 which is the predicted target of MO4. (d) Animals injected at the one-cell stage with the indicated morpholino were photographed at 48 hpi. At these doses of morpholinos a large number of animals develop normally and were used for regeneration studies. |