- Title
-
Unveiling the role of Ndrg1 gene on the oxidative stress induction behind the anticancer potential of styrylquinazoline derivatives
- Authors
- Malarz, K., Kuczak, M., Rurka, P., Rawicka, P., Boguszewska-Czubara, A., Jampilek, J., Mularski, J., Musiol, R., Mrozek-Wilczkiewicz, A.
- Source
- Full text @ Sci. Rep.
|
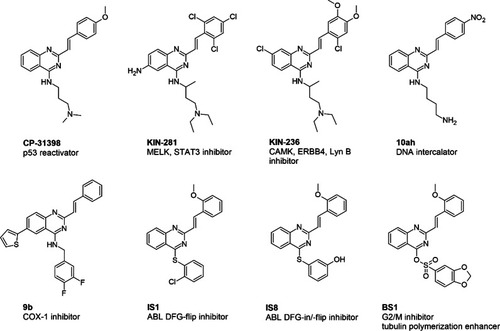
Diversity of 2-styrylquinazoline-core compounds and their biological signature in cancer cells. |
|
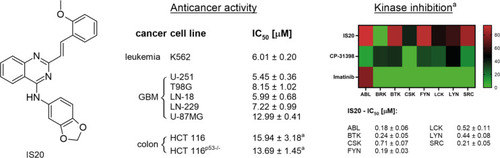
The chemical structure of IS20 (6b) styrylquinazoline, along with its anticancer activity on a panel of cancer cell lines with different |
|
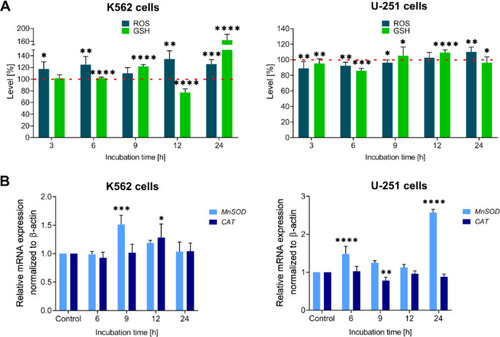
The time-dependent impact of the tested IS20 derivative on ROS (navy blue bars) and GSH (green bars) levels in the K562 and U-251 cells ( |
|
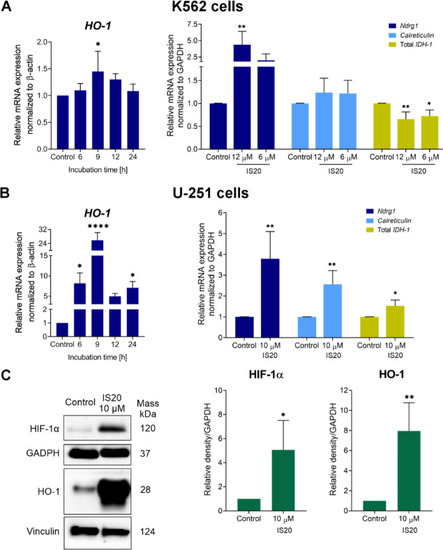
The impact of IS20 on the expression of selected genes and proteins associated with oxidative stress and iron regulation in the K562 ( |
|
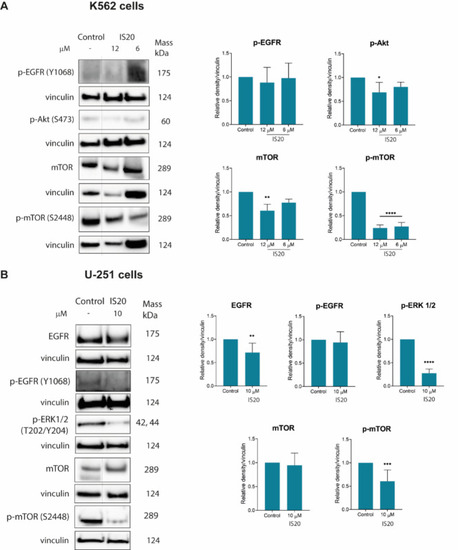
The impact of IS20 on the expression of EGFR and its downstream signaling proteins in K562 ( |
|
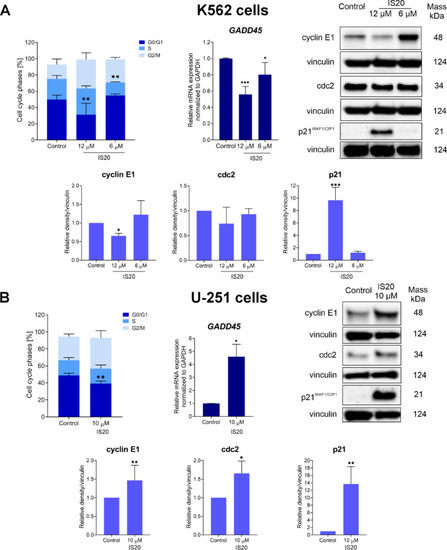
The impact of IS20 on the progression of the cell cycle and related molecules in the K562 ( |
|
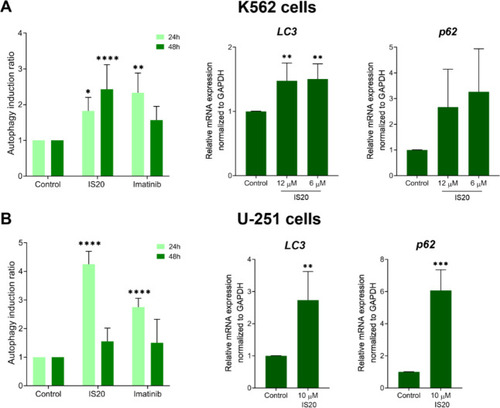
The influence of IS20 on the autophagy induction and expression of autophagy-related genes in the K562 ( |
|
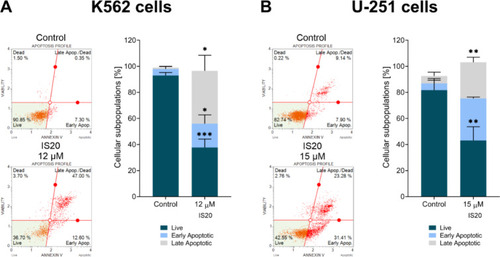
The impact of IS20 on the induction of apoptosis in the K562 ( |
|
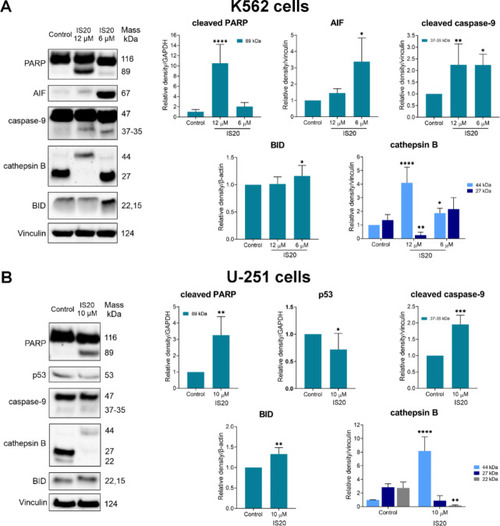
The impact of IS20 on the induction of expression of apoptosis-related proteins in the K562 ( |
|
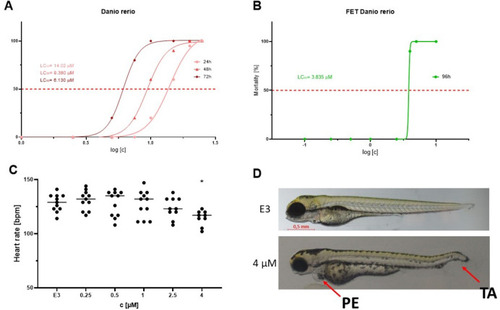
The toxicity of IS20 in |