|
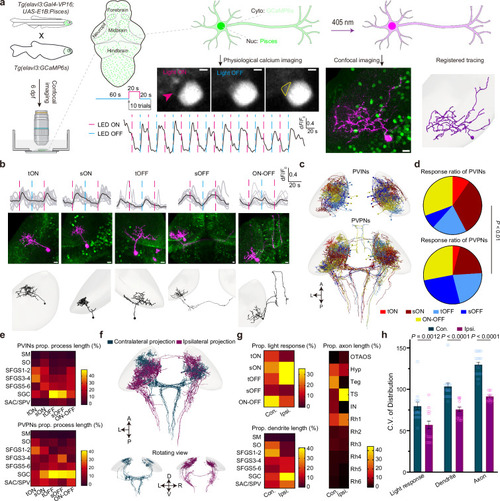
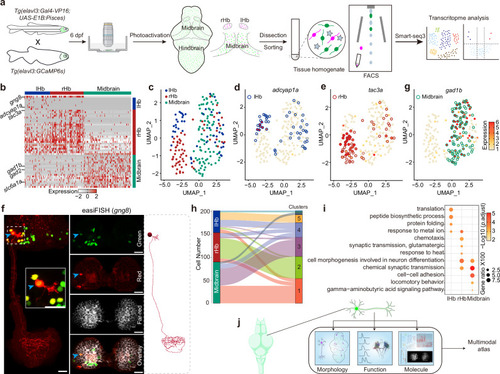
Transcriptome analysis of Pisces-activated GCaMP6s-positive neurons.a Overview of the workflow for isolating and analyzing single-cell transcriptomes from Pisces-activated, GCaMP6s-positive neurons in the left habenula (lHb), right habenula (rHb), and midbrain. b Heatmap showing differentially expressed genes across lHb, rHb, and midbrain neurons. Marker genes include gng8 (habenula-specific), adcyap1a (lHb), tac3a (rHb), gad1b, gad2, and slc6a1a (midbrain). c Uniform manifold approximation and projection (UMAP) clustering of neurons from lHb (n = 48), rHb (n = 64), and midbrain (n = 90) identify three distinct populations. Distribution patterns of habenular neurons expressing enriched marker genes in the UMAP clusters, including adcyap1a in the lHb (d) and tac3a in the rHb (e). f Representative z-axis maximum projection images of all 211 slices illustrating the integration of gng8 mRNA spatial localization and single habenular neuron morphology in zebrafish larvae expressing neuronal Pisces. Left: Morphology of individual habenular neurons, imaging before easiFISH with a zoomed-in view of the activated neuron (dashed square). Middle: Projection images of 10 slices showing gng8 mRNA expression detected by easiFISH. Nuclear-localized Pisces, activated neurons, and mRNA signals are shown in green, red, and far-red (white), respectively. After easiFISH, neurite signals diminish, leaving only nuclear fluorescence. Right: Morphological reconstructions of neurons before easiFISH. Activated neuron indicated by blue arrows. Similar results were observed from 3 larvae (ALE conditions). Scale bars: 20 μm. g Distribution patterns of midbrain neurons expressing enriched marker genes gad1b in the UMAP clusters. h Sankey plot illustrating the relationships between known lHb, rHb, and midbrain neurons and eight unsupervised cell clusters identified in the unbiased UMAP analysis. Clusters 1–5 are labeled; clusters 6–8 are included but unlabeled due to space limitations. Mutual information (MI = 0.50) indicates the distinct characteristics of each cell type. i Gene Ontology (GO) enrichment analysis for neurons from the three brain regions. Statistics were performed using a two-sided Hypergeometric test with Benjamini–Hochberg correction for multiple comparisons. Summary model integrating molecular, morphological, and functional data from single neurons across the whole brain in vivo using the Pisces system. See also Supplementary Fig. 7, Supplementary Movies 12 and 13.
|