- Title
-
Somites are a source of nephron progenitors in zebrafish
- Authors
- Peng, Z., Vanichapol, T., Nguyen, P.D., Chang, H.G., Kocha, K.M., O'Brien, L.L., Currie, P.D., Huang, P., Davidson, A.J.
- Source
- Full text @ Nat. Commun.
|
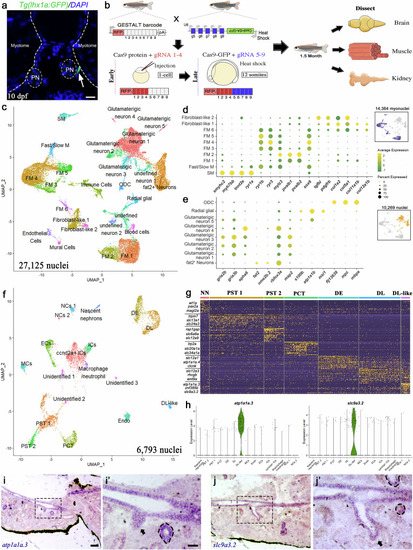
snRNA-Seq identifies various cell types from zebrafish muscle, kidney, and brain. |
|
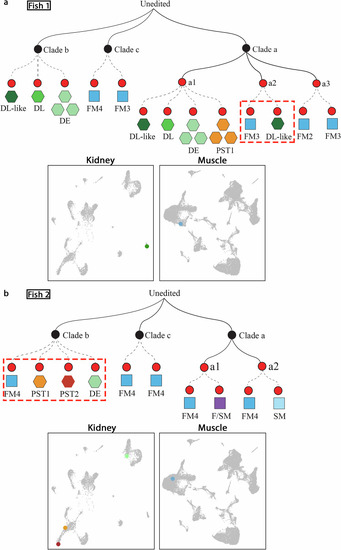
Reconstructed lineage trees reveal linkage between muscle and kidney cells in juvenile zebrafish. |
|
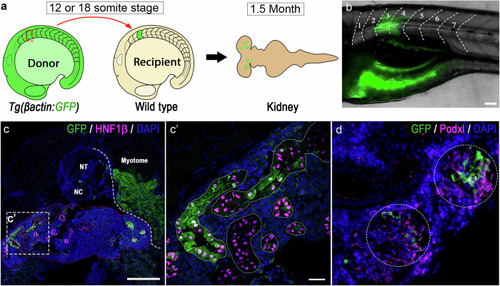
Transplanted GFP+ somite contributed to mesonephric nephrons in recipient fish. |
|
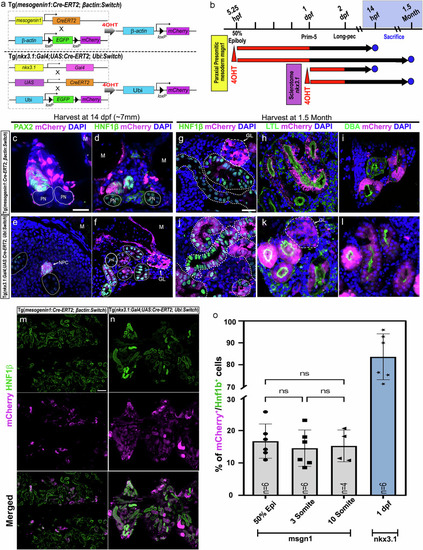
Cre-loxP based lineage-tracing techniques unveil a somitic origin for zebrafish mesonephric nephrons. |