- Title
-
Group B Streptococci lyse endothelial cells to infect the brain in a zebrafish meningitis model
- Authors
- Ravishankar, S., Tuohey, S.M., Ramos, N.O., Uchiyama, S., Hayes, M.I., Kang, K., Nizet, V., Madigan, C.A.
- Source
- Full text @ PLoS Biol.
|
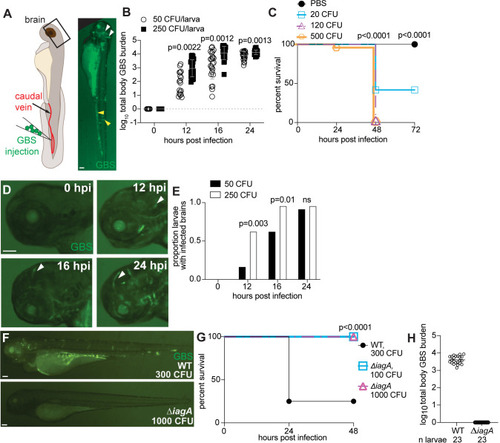
GBS infects the brain of zebrafish larvae in a time- and inoculum-dependent manner. |
|
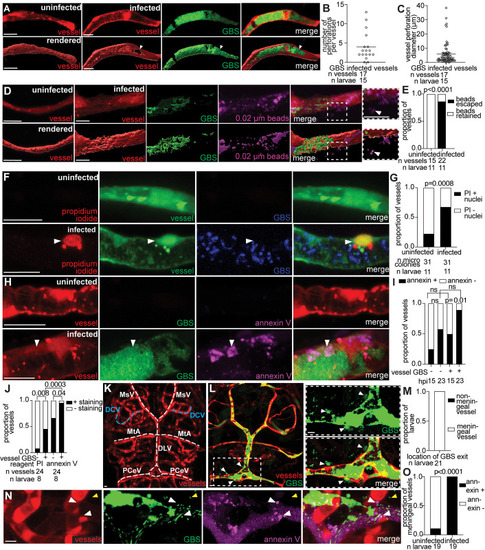
GBS does not use transcytosis or phagocytes to cross the BBB. |
|
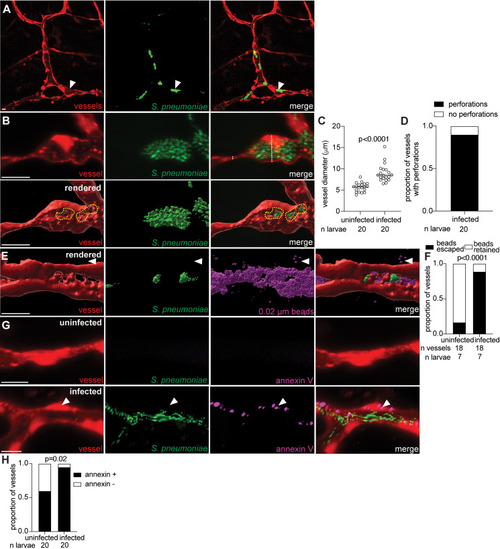
GBS lyses leptomeningeal endothelial cells to enter the brain. |
|
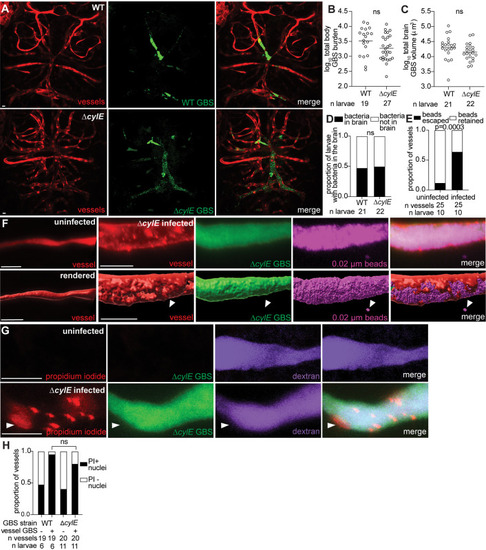
Endothelial cell lysis occurs independently of the primary GBS cytolysin, cylE. |
|
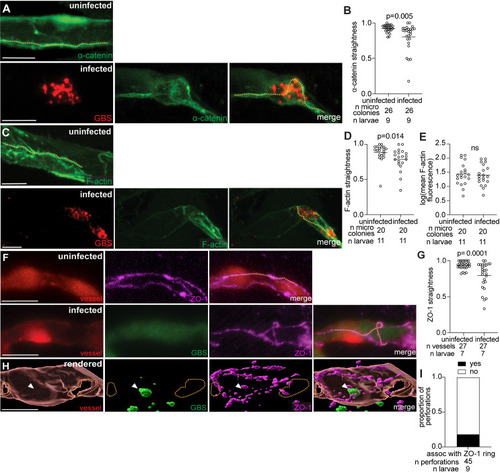
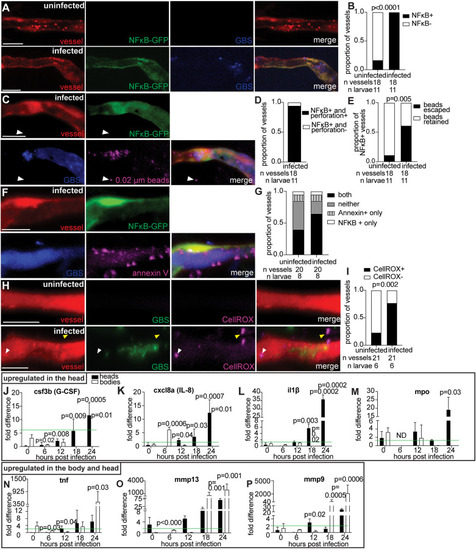
Blood–brain barrier tight and adherens junction proteins are likely disrupted during GBS infection due to endothelial cell lysis. |
|
Upregulation of proinflammatory markers suggests host responses contribute to GBS brain invasion. |
|
GBS microcolonies distort vessels and form obstructions. |
|
|