- Title
-
Characterization of a water soluble quininib prodrug that blocks metabolic activity and proliferation of multiple cancer cell lines
- Authors
- Tonelotto, V., Qaisar, A., McLoughlin, E.C., Cassaday, A., Kundu, K., Pendino, M., Marcone, S., O'Sullivan, J., Twamley, B., Jensen, L.D., Thorpe, S.D., Kennedy, B.N., O'Boyle, N.M.
- Source
- Full text @ Eur. J. Med. Chem.
|
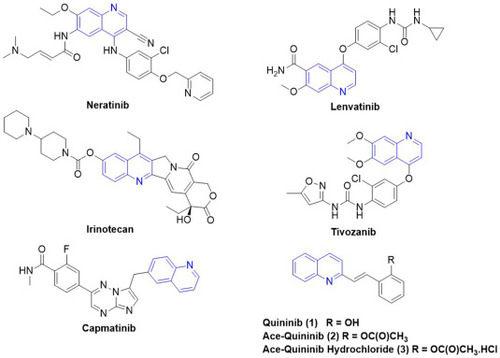
Structures of clinically approved anticancer quinoline-containing drugs, together with quininib and acetyl ester derivatives. |
|
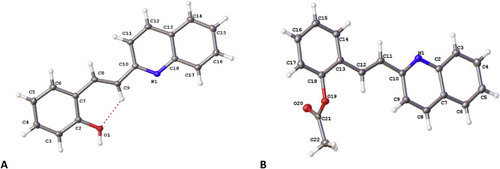
Molecular structure of (A) quininib (1) and (B) ace-quininib (2) with displacement parameters shown at 50 % probability. Intramolecular hydrogen bonding represented by dotted line. |
|
Stability of quininib and analogues at different pH values and in the presence of esterase. Stability of quininib (A), ace-quininib-HCl (3) (B) and ace-quininib (2) (C) at different pH values at room temperature (20 °C). Stability study (n = 2) was carried out in phosphate buffers at pH 4 (blue circles), 7.4 (red squares) or 9 (green triangles). Compound (1 mg/mL, 10 μL) was added to the phosphate buffer at room temperature. Samples were removed, vortexed and assayed at t = 0, 15, 30, 60, 120, 180, 240 and 1440 min. Compound stability was determined based on the percentage remaining at each timepoint compared to amount at t = 0 min. (D,E) Ace-quininib (2) is converted to quininib (1) in the presence of esterase enzymes. The conversion of ace-quininib (2) to quininib (1) was measured using a porcine esterase enzyme in PBS at pH 7.4. The reaction mixture was incubated in a water bath at 37 °C for 5 min. At t = 5 s, 30 s, 1 min, 2 min and 5 min, aliquots of 500 μL were removed and quenched with ice cold methanol: acetonitrile before analysis by HPLC. N = 3. (E) Retention time of quininib (1) is 4.9 min and ace-quininib (2) is 6.1 min. |
|
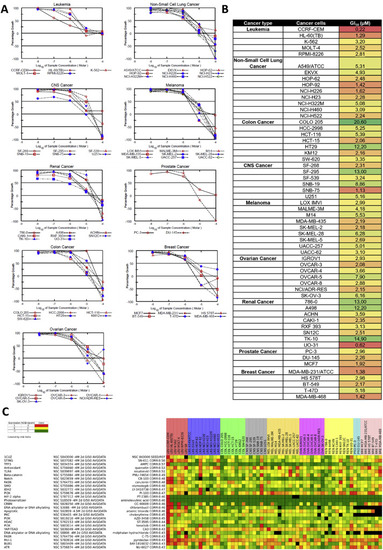
The anti-cancer effects of ace-quininib (2) in the NCI-60 human tumor cell line screen. (A) Each panel represents cell lines derived from of a particular type of cancer (left to right, top to bottom): 5 leukemia cancer cell lines, 9 non-small cell lung cancer (NSCLC) cell lines, 6 central nervous system (CNS) cell lines, 9 melanoma cell lines, 8 renal cancer cell lines, 2 prostate cancer cell lines, 7 colon cancer cell lines, 6 breast cancer cell lines, and 7 ovarian cancer cell lines. Ace-quininib (2) was tested at the concentration range of 0.01, 0.1, 1, 10, 100 μM and cells were exposed for 48 h. (B) Five-dose GI50 trend analysis of ace-quininib (2) against the full panel of human tumour cell lines. Red = most responsive cell lines, green = least responsive cell line. Values are shown in μM. (C) Heat map view of hierarchical clustering of NCI-60 growth response Pearson correlation patterns, derived from the COMPARE algorithm. The heat map ranks the entire database of tested compounds, in the order of the similarity of the GI50 response of the cell lines to the compounds in the database, to the responses of the cell lines to ace-quininib (2) (NSC: S843000). NCI-60 data for the targeted investigational oncology agents (IOA) set of 180 clinically approved agents and 820 investigational agents were used to run the COMPARE analysis. NCIEndpoint is color representation of unitDelta GI50 values ranging from 10−9 M (red) to 10−5 M (green). Cells are black when values were not obtained. |
|
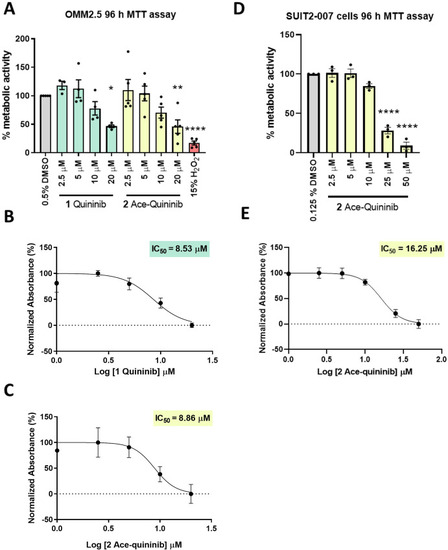
Ace-quininib (2) significantly reduces viability of OMM2.5 and SUIT2-007 cells. (A) A dose-dependent, significant decrease in OMM2.5 cell metabolic activity was observed following 96 h treatment with quininib (1) or ace-quininib (2) in comparison to 0.5 % DMSO treatment. One-way ANOVA with Dunnett's Test for Multiple Comparisons statistical analysis was performed; error bars represent mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. (N = 3–5). (B,C) IC50 values of quininib (1) (B) and ace-quininib (2) (C) in OMM2.5 cells following a 96 h treatment, as determined by metabolic activity assays. (D) A dose-dependent, significant decrease in SUIT2-007 cell metabolic activity was observed following 96 h treatment with ace-quininib (2) in comparison to 0.125 % DMSO treatment. One-way ANOVA with Dunnett's Test for Multiple Comparisons statistical analysis was performed; error bars represent mean ± SEM, ∗∗∗∗p < 0.0001 (N = 3). (E) IC50 value of ace-quininib (2) in SUIT2-007 cells following a 96 h treatment, as determined by metabolic activity assays. |
|
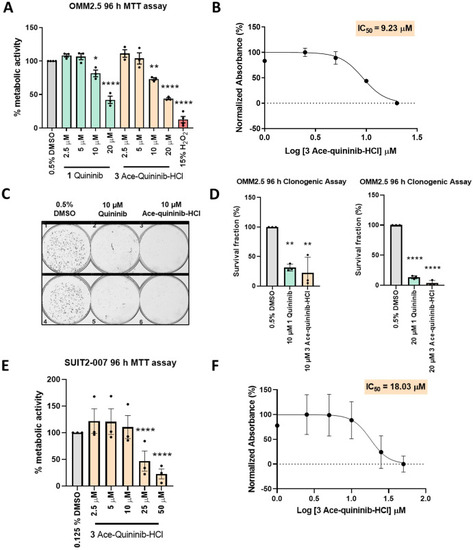
Quininib (1) and ace-quininib-HCl (3) significantly reduce viability of OMM2.5 and SUIT2-007 cells. (A) A dose-dependent, significant decrease in OMM2.5 cell metabolic activity was observed following 96 h treatment with quininib (1) or ace-quininib-HCl (3) in comparison to 0.5 % DMSO treatment. One-way ANOVA with Dunnett's Test for Multiple Comparisons statistical analysis was performed; error bars represent mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. (N = 3–5). (B) IC50 value of ace-quininib-HCl (3) in OMM2.5 cells following a 96 h treatment, as determined by metabolic activity assays. (C) Images of clones captured by GelCount™ system (Oxford Optronix) after 8 days of culture following treatment with DMSO control or 10 μM quininib (1) or 10 μM ace-quininib-HCl (3) for 96 h. Clones were stained with 0.5 % crystal violet before counting. 2000 cells were seeded and treated in duplicate in 6-well plates for each individual experiment and individual experiments were conducted three times. (D) Graphs show the percentage survival fraction of clones at 96 h post treatment with 10 μM quininib (1) or 10 μM ace-quininib-HCl (3) (left) and with 20 μM quininib (1) or 20 μM ace-quininib-HCl (3) (right). Statistical analysis was performed by ANOVA with Dunnett's post hoc multiple comparison test. Error bars are mean + SEM, ∗∗p < 0.01; ∗∗∗∗p < 0.0001. N = 3. (E) A dose-dependent, significant decrease in SUIT2-007 cell metabolic activity was observed following 96 h treatment with ace-quininib-HCl (3) in comparison to 0.125 % DMSO treatment. One-way ANOVA with Dunnett's Test for Multiple Comparisons statistical analysis was performed; error bars represent mean ± SEM, ∗∗∗∗p < 0.0001 (N = 3). (F) IC50 value of ace-quininib-HCl (3) in SUIT2-007 cells following a 96 h treatment, as determined by metabolic activity assays. |
|
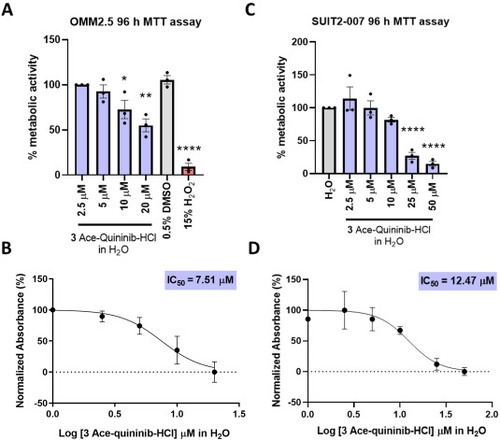
Water-soluble ace-quininib-HCl (3) significantly reduces viability of OMM2.5 and SUIT2-007. (A) A dose dependent decrease in cell metabolic activity was detected following 96 h of treatment with ace-quininib-HCl (3) solubilized in water. Statistical comparisons were performed between the lowest water-soluble drug concentration (2.5 μM) and both higher concentrations (5, 10, 20 μM) and 0.5 % DMSO, showing significant reductions in viability at higher doses. One-way ANOVA with Dunnett's Test for Multiple Comparisons statistical analysis was performed; error bars represent mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, (N = 3). (B) IC50 value of ace-quininib-HCl (3) in OMM2.5 cells following a 96 h treatment, as determined by metabolic activity assays. (C) A dose-dependent, significant decrease in SUIT2-007 cell metabolic activity was observed following 96 h treatment with ace-quininib-HCl (3) solubilized in water in comparison to 0.125 % water (control). One-way ANOVA with Dunnett's Test for Multiple Comparisons statistical analysis was performed; error bars represent mean ± SEM, ∗∗∗∗p < 0.0001 (N = 3). (D) IC50 value of ace-quininib-HCl (3) solubilized in water in SUIT2-007 cells following a 96 h treatment, as determined by metabolic activity assays. |
|
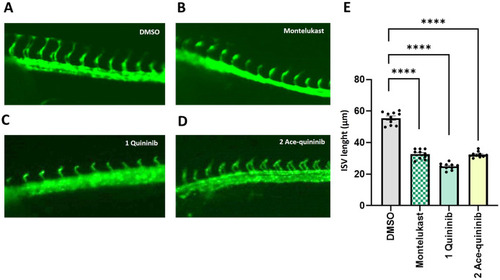
Quininib (1) and ace-quininib (2) inhibit angiogenesis in zebrafish. (A–D) Live fluorescence images of ISVs of zebrafish embryos (24 hpf) treated for 20 h with (A) DMSO 0.1 % v/v; (B) montelukast (12.5 μM), (C) quininib (1) (12.5 μM), (D) ace-quininib (2) (12.5 μM). (E) Quantification of the average length of ISV of zebrafish embryos. Embryos were treated with DMSO (0.1 % v/v), montelukast (12.5 μM), quininib (1) (12.5 μM), and ace-quininib (2) (12.5 μM). N = 10. One-way analysis of variance (ANOVA) was performed for statistical analysis. Error bars represent mean ± SEM, ∗∗∗∗p < 0.0001. N = 10. |