- Title
-
Tie2 activity in cancer associated myofibroblasts serves as novel target against reprogramming of cancer cells to embryonic-like cell state and associated poor prognosis in oral carcinoma patients
- Authors
- Mitra, P., Saha, U., Stephen, K.J., Prasad, P., Jena, S., Patel, A.K., Bv, H., Mondal, S.K., Kurkalang, S., Roy, S., Ghosh, A., Roy, S.S., Das Sarma, J., Biswas, N.K., Acharya, M., Sharan, R., Arun, P., Jolly, M.K., Maitra, A., Singh, S.
- Source
- Full text @ J. Exp. Clin. Cancer Res.
|
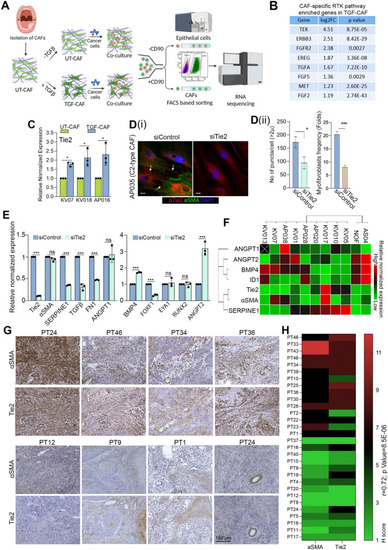
C2-CAF expressed higher levels of Tie2 and positively correlated with αSMA-high stromal fibroblasts in primary tumors.( |
|
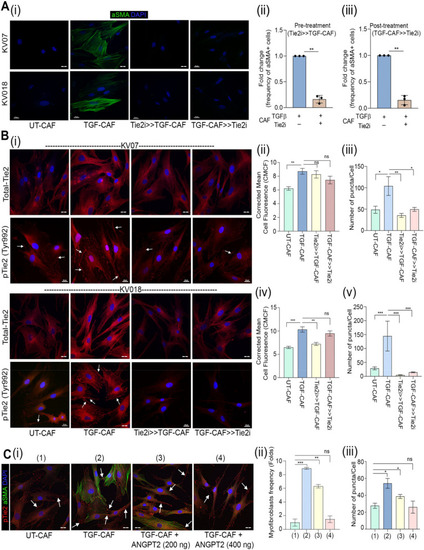
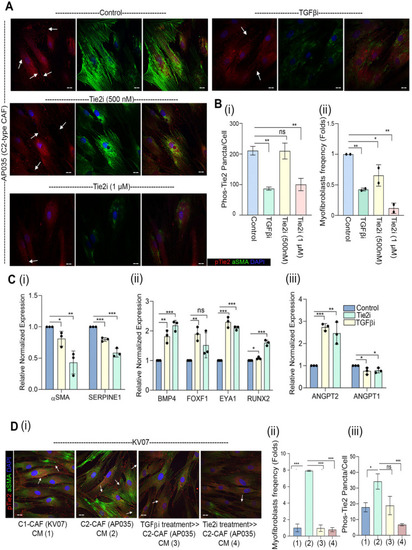
Tie2 plays essential role in induction as well as sustenance of TGFβ-induced myofibroblastic differentiation of CAF. ( |
|
Tie2-activity is regulated in an autocrine manner.( |
|
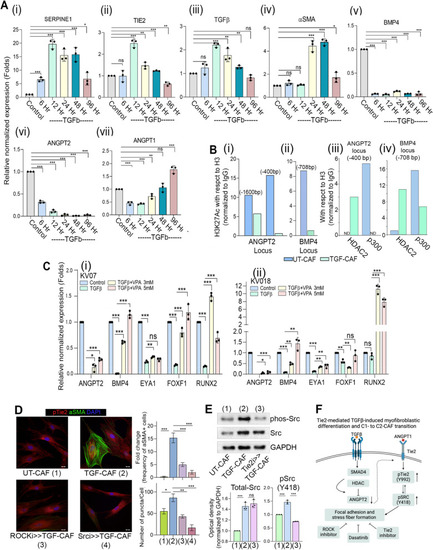
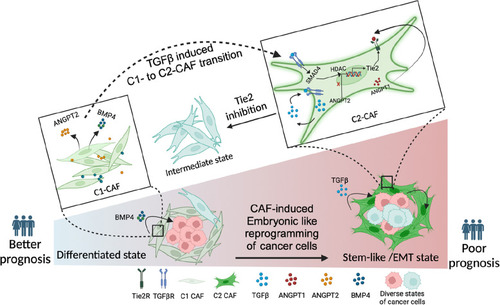
TGFβ-induced histone deacetylation drives transcriptional state changes associated with transition of C1- to C2-CAF.( |
|
Endogenous-TGFβ is necessary and sufficient in driving Tie2-ANGPT signaling.( |
|
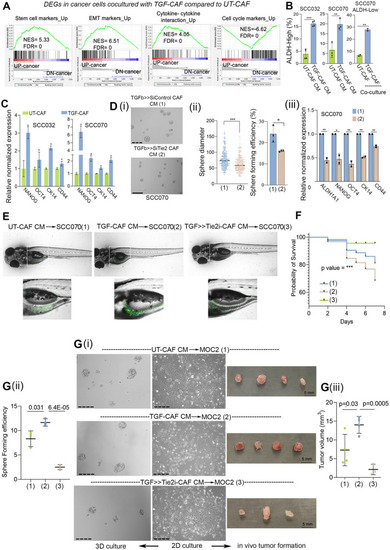
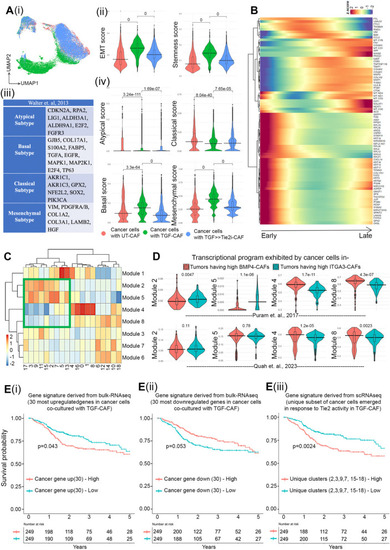
TGFβ-induced myofibroblastic C2-CAF reprograms oral-cancer cells to acquire embryonic-like transcriptome state.( |
|
CAF-specific Tie2 regulates cancer cell plasticity and stemness in oral cancer cells.( |
|
Tie2 responsive single cell gene expression data derived modules translate to clinical output of HNSCC patients.( |
|
CAF-specific Tie2 activity in reprogramming of oral cancer cells. We have previously identified and characterized C1-CAF and C2-CAF in oral cancer. C1-CAFs exhibit higher-BMP4 expression, whereas C2-CAF exhibit myofibroblastic phenotype with aSMA-positive stress fiber formation. The C2-CAFs supported stem-like properties in cancer cells. Here, we have explored the possible mechanism and demonstrated that the TGFβ-induced myofibroblastic differentiation and conversion of C1-CAF into C2-CAF is mediated through the activation of Tie2-signaling with suppression of its antagonist-ANGPT2 due to HDAC-mediated deacetylation of its promoter. Furthermore, Tie2-inhibition was found to convert TGFβ-induced-CAF towards the transcriptional state of C1-CAF. Functionally, TGFβ-induced CAF reprogramed oral cancer cells into embryonic-like state with enhanced stemness and EMT properties. Emphasizing its clinical translational value, the specific gene-signature derived from the cancer cells, reprogrammed by TGFβ-induced Tie2-activated-CAF, may predict the poor prognosis in head and neck cancer patients |