- Title
-
A sensitive soma-localized red fluorescent calcium indicator for in vivo imaging of neuronal populations at single-cell resolution
- Authors
- Zhou, S., Zhu, Q., Eom, M., Fang, S., Subach, O.M., Ran, C., Alvarado, J.S., Sunkavalli, P.S., Dong, Y., Wang, Y., Hu, J., Zhang, H., Wang, Z., Sun, X., Yang, T., Mu, Y., Yoon, Y.G., Guo, Z.V., Subach, F.V., Piatkevich, K.D.
- Source
- Full text @ PLoS Biol.
|
Design of FRCaMPi and characterization in HeLa cells. |
|
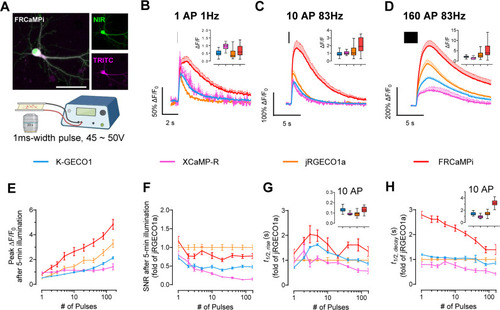
Characterization of FRCaMPi in primary mouse hippocampal neurons. |
|
SomaFRCaMPi engineering and characterization via electric field stimulation in primary mouse hippocampal neurons. |
|
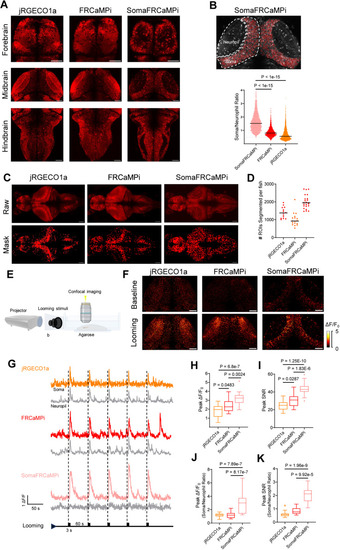
Expression and performance of jRGECO1a, FRCaMPi, and SomaFRCaMPi in reporting neuronal activities in zebrafish larvae. |
|
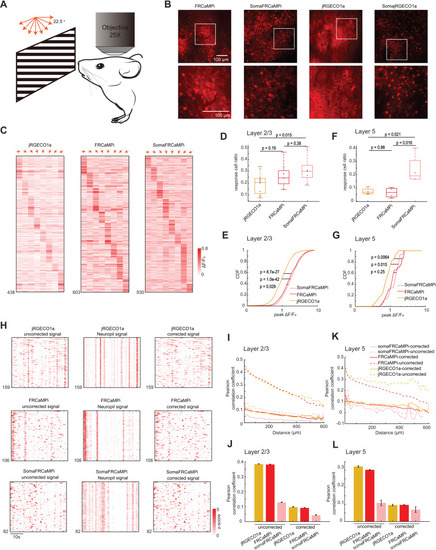
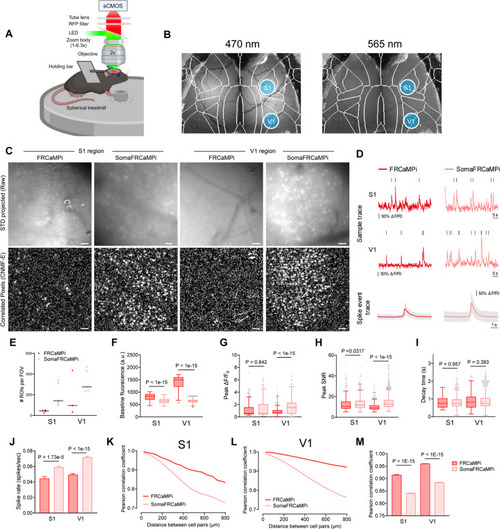
In vivo neural population imaging in mice V1 cortex. |
|
Dual-color in vivo calcium imaging of NTS tuning during gastric distension. |
|
Recording of FRCaMPi and SomaFRCaMPi dynamics in |