- Title
-
Chr23-miR-200s and Dmrt1 Control Sexually Dimorphic Trade-Off Between Reproduction and Growth in Zebrafish
- Authors
- Ge, S., Liu, Y., Huang, H., Yu, J., Li, X., Lin, Q., Huang, P., Mei, J.
- Source
- Full text @ Int. J. Mol. Sci.
|
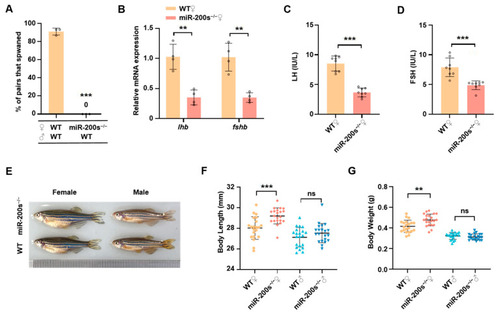
Impaired reproduction and promoted growth in female chr23-miR-200s-KO zebrafish. ( |
|
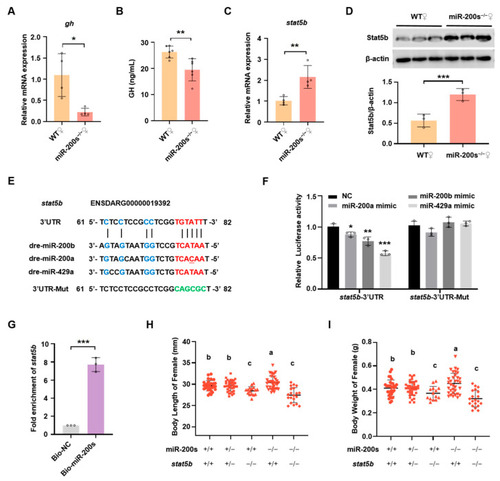
Chr23-miR-200s regulate growth of female zebrafish through targeting |
|
|
|
|
|
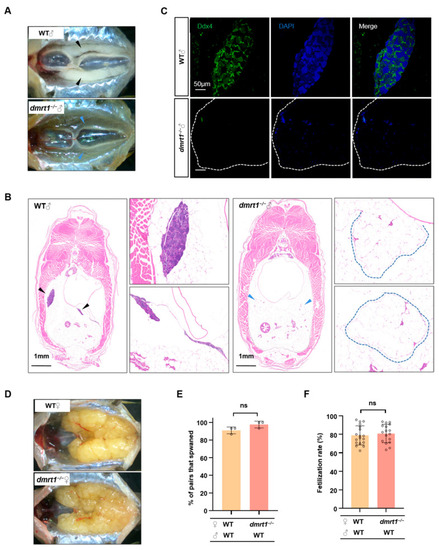
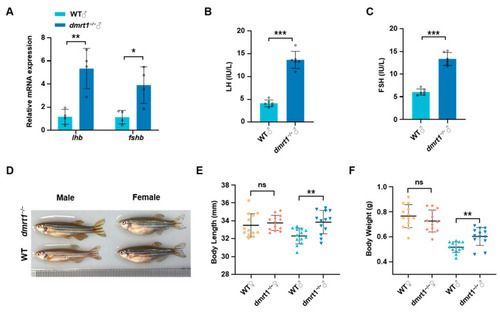
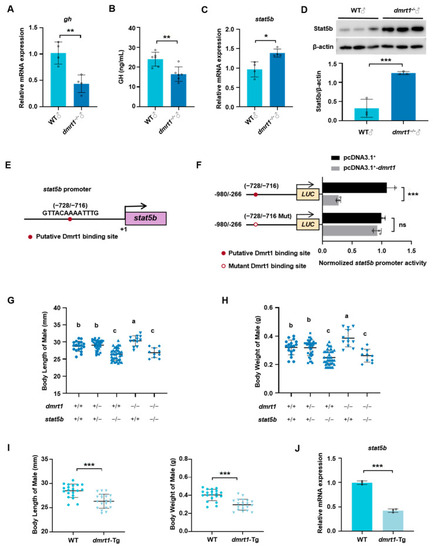
Dmrt1 regulates somatic growth in male zebrafish by mediating |