- Title
-
The transmembrane glycoprotein Gpnmb is required for the immune and fibrotic responses during zebrafish heart regeneration
- Authors
- Gupta, S., Bajwa, G.K., El-Sammak, H., Mattonet, K., Günther, S., Looso, M., Stainier, D.Y.R., Marín-Juez, R.
- Source
- Full text @ Dev. Biol.
|
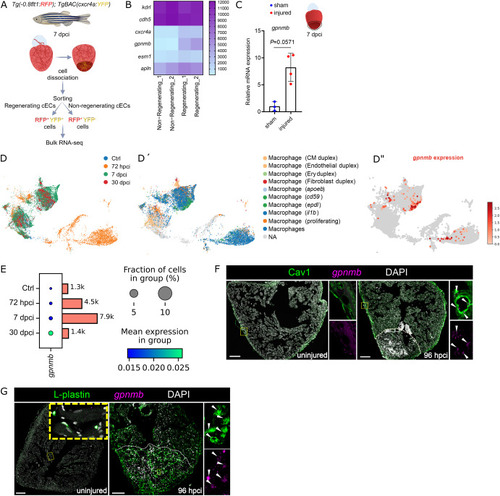
gpnmb is upregulated in regenerating coronary endothelial cells and macrophages after cardiac cryoinjury in zebrafish. A. Experimental plan for the bulk RNA-seq analysis on sorted regenerating (-0.8 flt1:RFP+/cxcr4a:YFP+) and non-regenerating (-0.8 flt1:RFP+/cxcr4a:YFP−) coronary endothelial cells from Tg(-0.8flt1:RFP); TgBAC(cxcr4a:YFP) whole ventricles at 7 dpci. B. Heatmap showing endothelial marker genes (kdrl, cdh5, crcr4a, esm1, and apln) and candidate genes (gpnmb and plxdc2) in the regenerating and non-regenerating coronary endothelial populations. C. RT-qPCR analysis of gpnmb mRNA levels at 7 dpci in the injured tissue relative to sham-operated hearts; Ct values listed in Supplementary Table S1. D-E. Reanalysis of the published single-cell RNA sequencing (scRNA-seq) dataset GSE159032 (Hu et al., 2022); UMAP representation of the different macrophage clusters highlighting 1) their distribution at several time-points (D), 2) their identity (D′), and 3) gpnmb expression (D′′); dot plots of gpnmb expression in macrophages in the adult zebrafish heart (E). F-G. in situ hybridization chain reaction (HCR) for gpnmb expression and immunostaining for Cav1 (F) and L-plastin (G) on sections of uninjured and 96 hpci ventricles. Scale bars: 100 μm. |
|
gpnmb mutants exhibit altered pro- and anti-inflammatory responses after cardiac cryoinjury in zebrafish. A. Illustration of gpnmbbns595 full locus deletion allele generated using the CRISPR/Cas9 technology. RT-qPCR analysis of gpnmb mRNA levels in 5 dpf larvae, dissected caudal fins from adult zebrafish, and 7 dpci injured tissue from adult ventricles, comparing gpnmb+/+ and gpnmb−/−siblings; Ct values listed in Supplementary Table S1; ND= Not detectable. B. Immunostaining for EGFP (macrophages, magenta) with DAPI (DNA marker, blue) counterstaining on sections of Tg(mpeg1:EGFP); gpnmb+/+ and Tg(mpeg1:EGFP); gpnmb−/− cryoinjured ventricles at 7 dpci. C. mpeg1:EGFP+ cell number in gpnmb+/+ and gpnmb−/− BZI tissue (100 μm) at 7 dpci. D. Immunostaining for Mpx (neutrophils, white) with DAPI (DNA marker, blue) counterstaining on sections of gpnmb+/+ and gpnmb−/− cryoinjured ventricles at 96 hpci. E. Mpx+ cell number in gpnmb+/+ and gpnmb−/− injured tissues at 96 hpci. F. Experimental plan for the bulk RNA-seq analysis performed on the BZI tissue of gpnmb−/− and gpnmb+/+ cryoinjured ventricles at 96 hpci. G. Heatmap showing differentially expressed genes of interest in the BZI tissue of gpnmb−/− and gpnmb+/+ cryoinjured ventricles at 96 hpci. H. GSE analysis plot of TNF-α signaling via NF-kB, the complement system and antigen response from the 96 hpci gpnmb−/− vs gpnmb+/+ transcriptomic analysis. White dotted lines delineate the injured area; white arrowheads point to mpeg1:EGFP+ (B) and Mpx+ (D) cells. Statistical test: Student's t-test (C, E). Scale bars: 100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.) |
|
gpnmb mutants exhibit reduced fibroblast activation after cardiac cryoinjury in zebrafish. A. GSE analysis plot of collagen formation, biosynthesis, and assembly from the 96 hpci gpnmb−/− vs gpnmb+/+ cryoinjured ventricles transcriptomic analysis. B. Heatmap showing differentially expressed genes of interest in the BZI tissue of gpnmb−/− and gpnmb+/+ cryoinjured ventricles at 96 hpci. C. RT-qPCR analysis of acta2 mRNA levels in the gpnmb−/− injured tissue normalized to gpnmb+/+ injured tissue at 7 dpci; Ct values listed in Supplementary Table S1. D. Immunostaining for αSMA (myofibroblasts, white) with DAPI (DNA marker, blue) counterstaining on sections of gpnmb+/+ and gpnmb−/− cryoinjured ventricles at 7 dpci. E. αSMA+ cell numbers in gpnmb+/+ and gpnmb−/− injured tissues at 7 dpci. F. AFOG staining on sections of gpnmb+/+ and gpnmb−/− cryoinjured ventricles at 30 dpci. G. AFOG staining on sections of gpnmb+/+ and gpnmb−/− cryoinjured ventricles at 90 dpci. H. Graphs showing the representation of groups (y-axis) of different scar area sizes (different colors) at 30 and 90 dpci for gpnmb+/+ and gpnmb−/− cryoinjured ventricles. Mean scar area sizes for gpnmb+/+ and gpnmb−/− at 30 dpci are 9.4 and 11.6 % respectively. Mean scar area sizes for gpnmb+/+ and gpnmb−/− at 90 dpci are 0.9 and 1.6 % respectively. White dotted lines delineate the injured area; white arrowheads point to αSMA+ cells (E). Statistical tests: Non-parametric Mann-Whitney test (C), Student's t-test (E). Scale bars: 100 μm (D) and 200 μm (F and H). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.) |
|
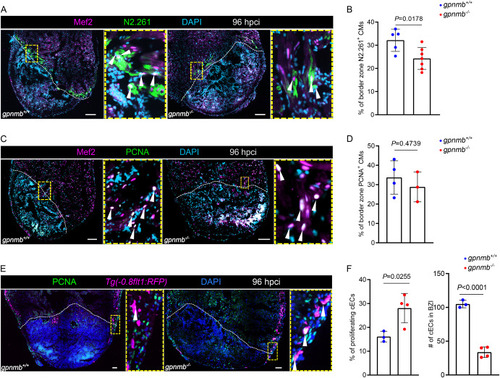
gpnmb mutants exhibit reduced cardiomyocyte dedifferentiation and reduced coronary endothelial cell numbers after cardiac cryoinjury in zebrafish. A. Immunostaining for Mef2 (cardiomyocyte nuclei, green), N2.261 (embryonic myosin heavy chain, magenta) with DAPI (DNA marker, cyan) counterstaining on sections of gpnmb+/+ and gpnmb−/− cryoinjured ventricles at 96 hpci. B. Quantification of dedifferentiating cardiomyocytes in border zone areas (100 μm) at 96 hpci. C. Immunostaining for Mef2 (cardiomyocyte nuclei, green), PCNA (proliferation marker, magenta) with DAPI (DNA marker, cyan) counterstaining on sections of gpnmb+/+ and gpnmb−/− cryoinjured ventricles at 96 hpci. D. Quantification of proliferating cardiomyocytes in border zone areas (100 μm) at 96 hpci. E. Immunostaining for PCNA (proliferation marker, green), RFP (coronary endothelial cells, magenta) with DAPI (DNA marker, blue) counterstaining on sections of Tg(-0.8flt1:RFP); gpnmb+/+ and Tg(-0.8flt1:RFP); gpnmb−/− cryoinjured ventricles at 96 hpci. F. Quantification of proliferating and total number of coronary endothelial cells in the BZI tissue (200 μm) at 96 hpci. White dotted lines delineate the injured area; white arrowheads point to dedifferentiating cardiomyocytes (A), proliferating cardiomyocytes (C), and proliferating coronary endothelial cells (E). Statistical test: Student's t-test (B, D, F). Scale bars: 100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.) PHENOTYPE:
|
Reprinted from Developmental Biology, , Gupta, S., Bajwa, G.K., El-Sammak, H., Mattonet, K., Günther, S., Looso, M., Stainier, D.Y.R., Marín-Juez, R., The transmembrane glycoprotein Gpnmb is required for the immune and fibrotic responses during zebrafish heart regeneration, , Copyright (2025) with permission from Elsevier. Full text @ Dev. Biol.