- Title
-
Understanding the Role of Exercise and Probiotic Interventions on Non-Alcoholic Fatty Liver Disease Alleviation in Zebrafish: Dialogue Between the Gut and Liver
- Authors
- Gu, X., Yuan, L., Gan, L., Zhang, Z., Zhou, S., Fu, Z., Liu, Y., Xin, Z., Cheng, S., Zhou, X., Yan, H., Wang, Q.
- Source
- Full text @ Int. J. Mol. Sci.
|
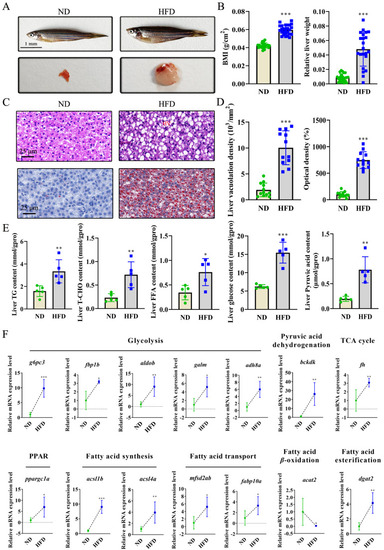
High-fat diet (HFD)-induced non-alcoholic fatty liver disease (NAFLD) progression involved in glucose and fatty acid metabolism: ( |
|
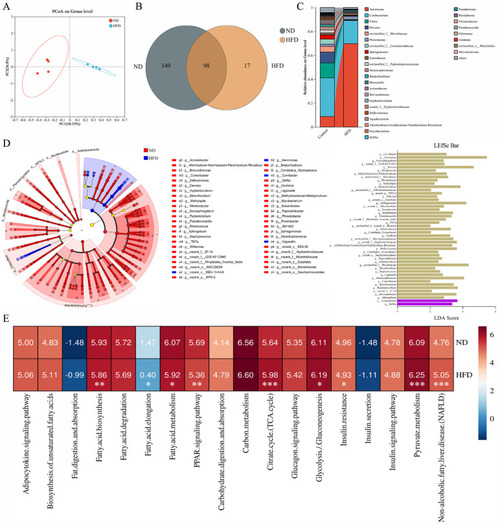
Alterations in gut microbiota may induce NAFLD by changing the glucose and fatty acid metabolism: ( |
|
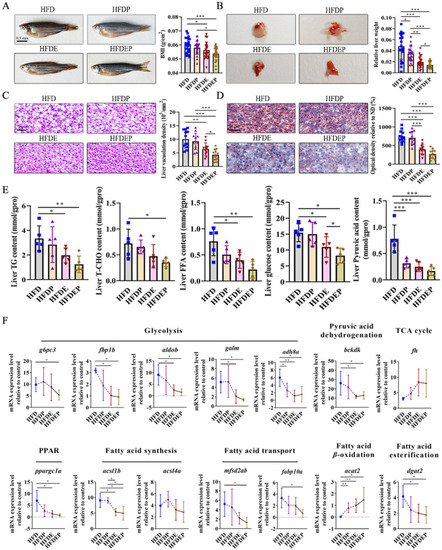
Effects of different intervention strategies on NAFLD: ( |
|
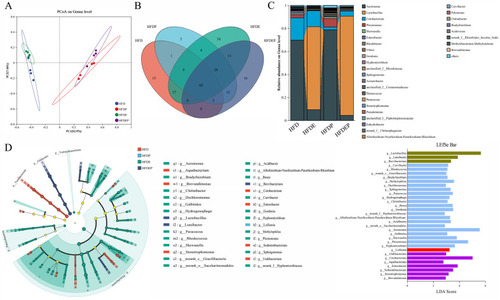
Gut microbial change under different intervention modes: ( |
|
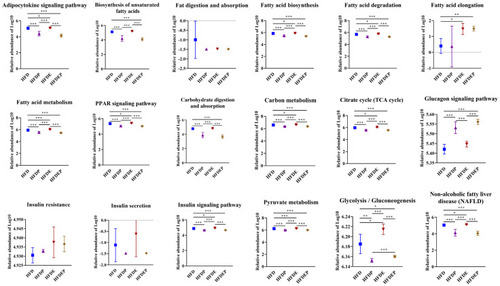
Gut microbial functional change by different intervention modes. Comparison of biological processes associated with glucose and fatty acid metabolism in different groups based on log10 of KEGG pathway abundances at level 3 using PICRUSt2. Values are the mean ± SD. * |
|
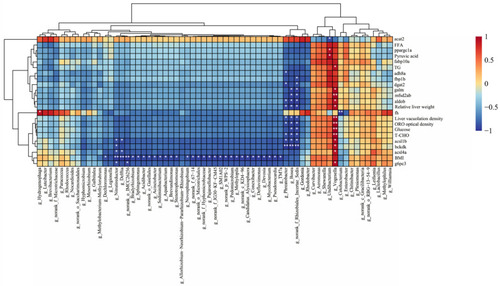
Correlation analysis between altered gut microbiota and lipid indicators. * |
|
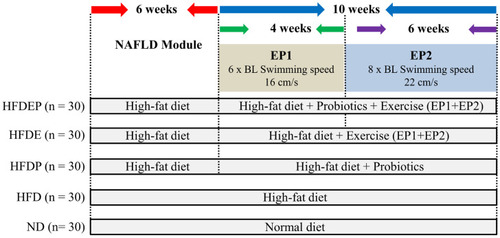
Schematic of experimental design. |