- Title
-
Dynamic interplay of cNHEJ and MMEJ pathways of DNA double-strand break repair during embryonic development in zebrafish
- Authors
- Carrara, M., Gaillard, A.L., Brion, A., Duvernois-Berthet, E., Giovannangeli, C., Concordet, J.P., Pézeron, G.
- Source
- Full text @ Sci. Rep.
|
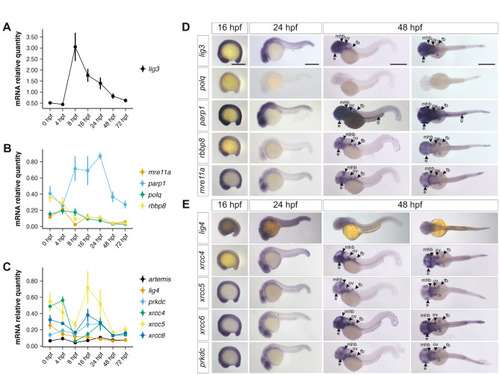
Analysis of MMEJ- and cNHEJ-related transcripts levels and distribution throughout embryonic development. ( |
|
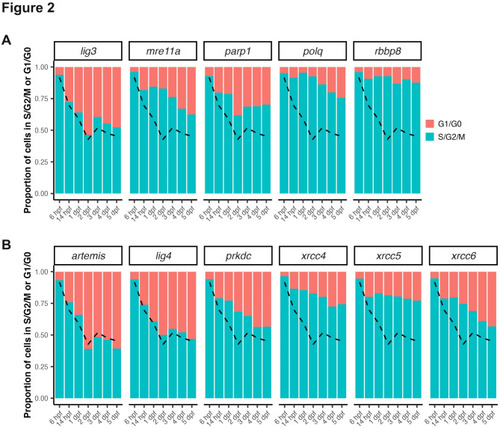
Analysis of mitotic activity in cells expressing MMEJ- and cNHEJ- related genes during embryogenesis. ( |
|
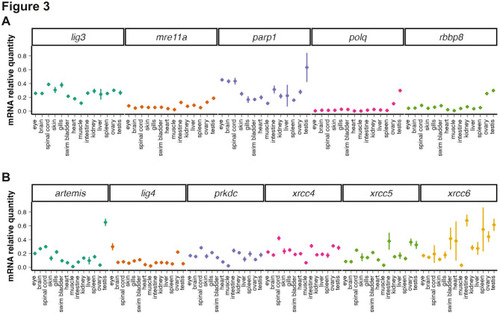
Analysis of mRNA levels of MMEJ- and cNHEJ- related genes across adult tissues. ( |
|
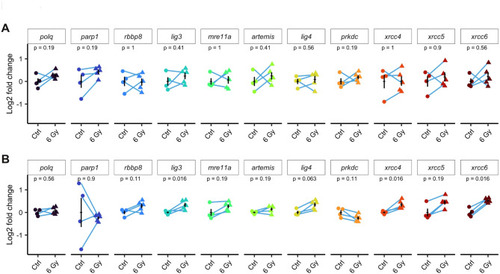
Analysis of mRNA levels of MMEJ- and cNHEJ-related genes in response to ionizing radiation exposure in embryos. ( |
|
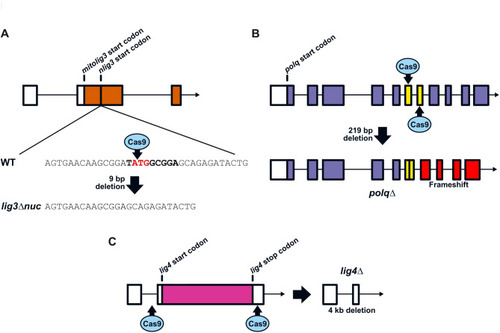
Inactivation of MMEJ and cNHEJ DNA repair pathways through CRISPR/Cas9-mediated gene knockout. ( |
|
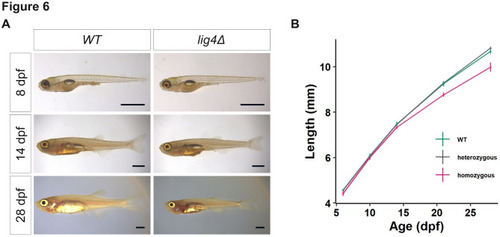
DNA ligase 4 is required for larval growth. ( |
|
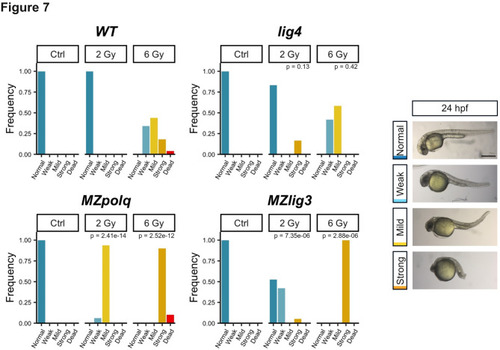
Inactivation of MMEJ sensitizes zebrafish embryos to IR. Embryos at 4 hpf were exposed to ionizing radiation (0, 2 and 6 Gy) and allowed to develop until 24 hpf. Phenotype was scored at 24 hpf according to the right panel scale. Weak = minor deformation of the tail, Mild = major deformation of the tail, Strong = atrophied tail and head, dead embryos were not imaged. WT, |
|
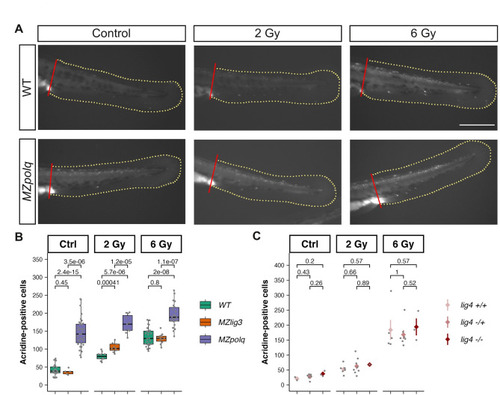
Cell death following irradiation in MMEJ and cNHEJ mutants. 24-hpf embryo from wildtype, |
|
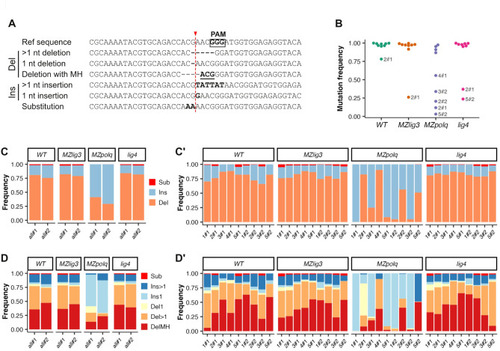
NGS analysis of the mutation spectrum after CRISPR/Cas9-mediated mutagenesis. CRISPR/Cas9-mediated mutagenesis was performed at 10 loci by injecting independently two mixes (#1 and #2) of five Cas9-GFP RNPs into zygotes (1–2 cell stage). For each mix, DNA was extracted at 9 hpf from pools of 20 embryos and PCR amplicons from all loci were sequenced, excepted one that could not be amplified. ( |