- Title
-
Deletion of ddx4 Ovary-Specific Transcript Causes Dysfunction of Meiosis and Derepress of DNA Transposons in Zebrafish Ovaries
- Authors
- Chen, Y., Lin, X., Dai, J., Bai, Y., Liu, F., Luo, D.
- Source
- Full text @ Biology (Basel)
|
|
|
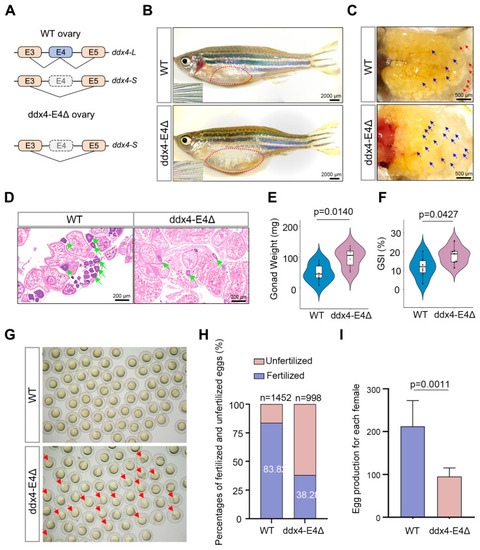
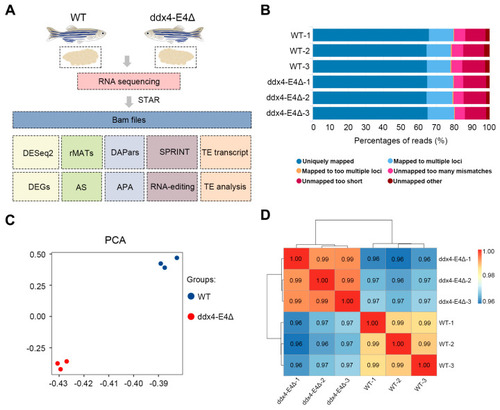
RNA sequencing of WT and ddx4-E4Δ zebrafish ovaries. ( |
|
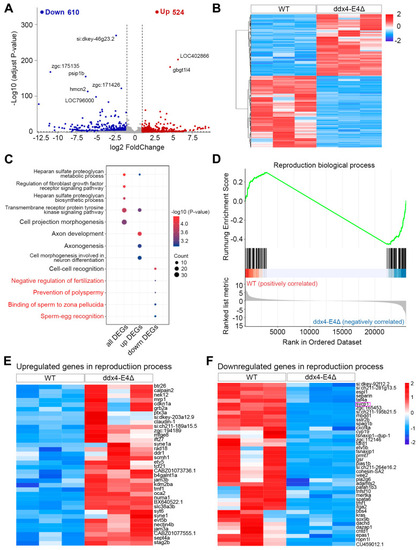
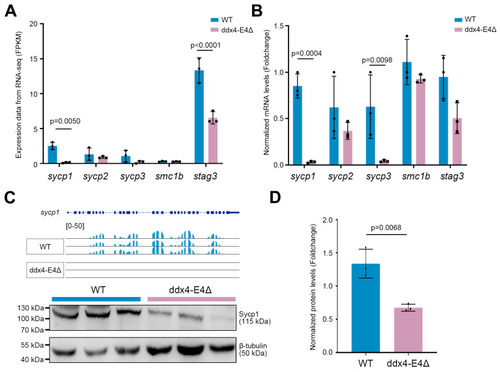
Identification and functional enrichment analysis of DEGs between WT and ddx4-E4Δ zebrafish ovaries. ( |
|
|
|
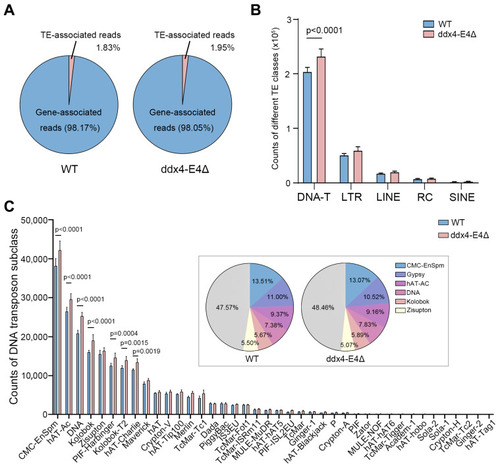
Identification of TEs and quantitative analysis of TE expression in WT and ddx4-E4Δ zebrafish ovaries. ( |
|
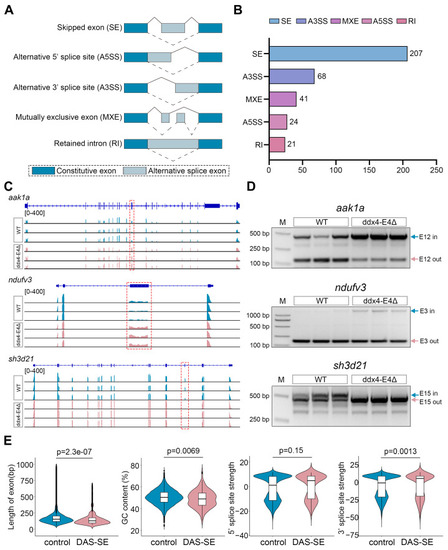
Identification of differential alternative splicing events between WT and ddx4-E4Δ zebrafish ovaries. ( |
|
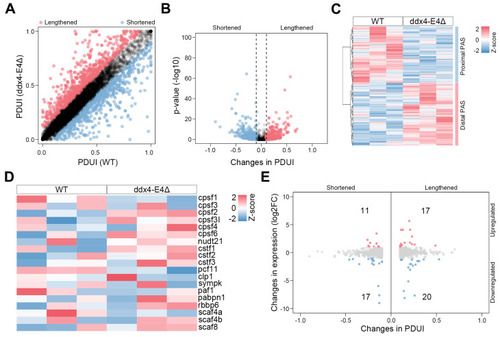
Identification of APA events between WT and ddx4-E4Δ zebrafish ovaries. ( |
|
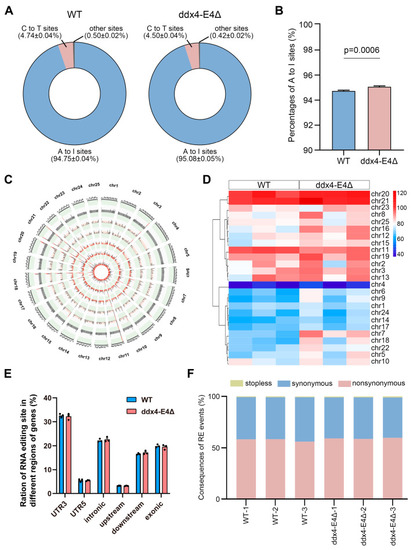
Identification of RNA editing events in WT and ddx4-E4Δ zebrafish ovaries. ( |