- Title
-
Analysis of multi-condition single-cell data with latent embedding multivariate regression
- Authors
- Ahlmann-Eltze, C., Huber, W.
- Source
- Full text @ Nat. Genet.
|
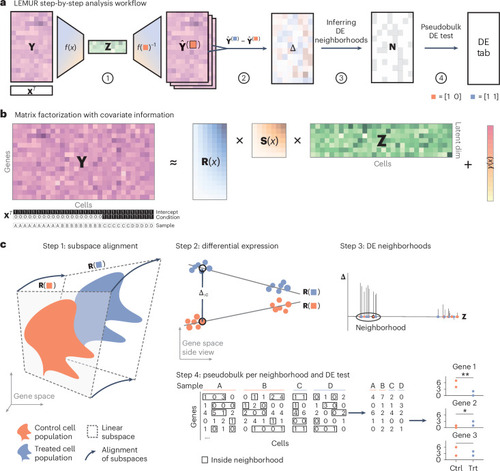
Conceptual overview of LEMUR. |
|
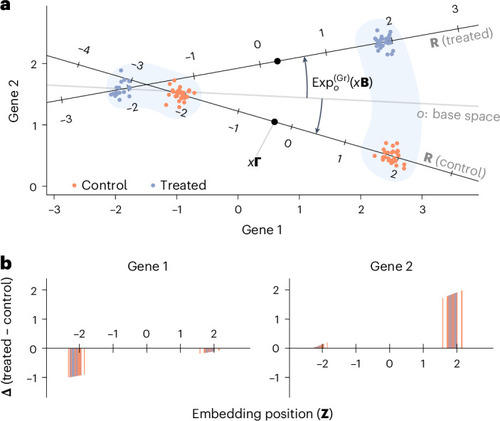
Stylized example with two genes observed in two groups of cells in two conditions. |
|
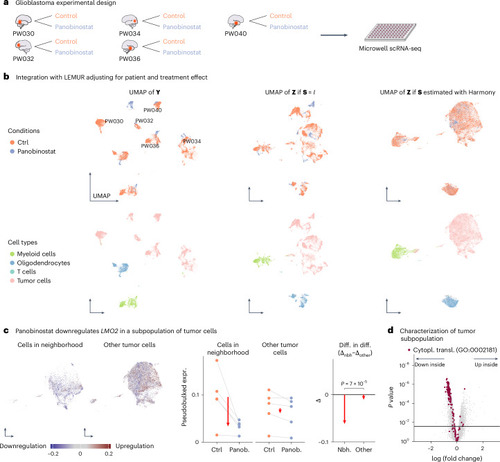
Analysis of five glioblastoma tumor biopsies. |
|
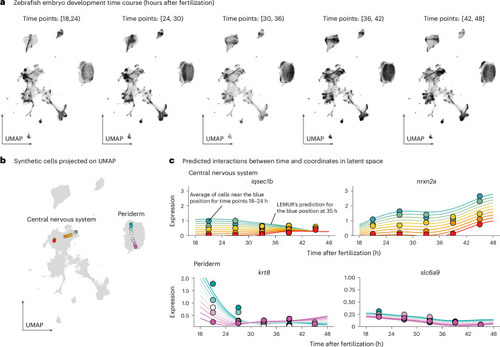
Analysis of a time course single-cell experiment. |
|
Analysis of a spatial single-cell experiment. |