Fig. 1
- ID
- ZDB-FIG-250318-17
- Publication
- Ahlmann-Eltze et al., 2025 - Analysis of multi-condition single-cell data with latent embedding multivariate regression
- Other Figures
- All Figure Page
- Back to All Figure Page
|
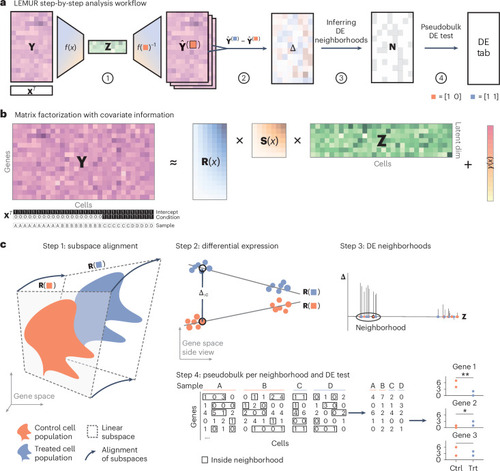
Conceptual overview of LEMUR. |