- Title
-
Integrated Studies on Male Reproductive Toxicity of Decabromodiphenyl Ethane in Zebrafish Spermatozoa Ex Vivo, Male Zebrafish in Vivo, and GC-1 Cells in Vitro
- Authors
- Yang, L., Zhang, Y., Hua, J., Song, G., Li, F., Zheng, N., Zhang, T., Xu, Z., Ren, X., Zhu, B., Han, Y., Guo, Y., Han, J., Zhou, B.
- Source
- Full text @ Environ. Health Perspect.
|
Effects of DBDPE |
|
Effects of DBDPE |
|
Histological observations of male zebrafish testes after 2-month |
|
Whole-proteome and phosphoproteome analysis of male zebrafish testes after |
|
DNA damage in zebrafish testes exposed to DBDPE |
|
Germ cell apoptosis and expression of proteins related to cell apoptosis in zebrafish testes exposed to DBDPE |
|
Energy metabolic level in zebrafish testes exposed to DBDPE |
|
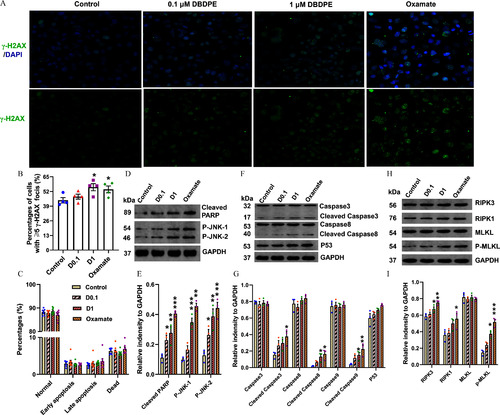
DNA damage and expression of proteins related to apoptosis or necroptosis in mouse spermatogonial GC-1 cells exposed to DBDPE and oxamate (LDH inhibitor) |
|
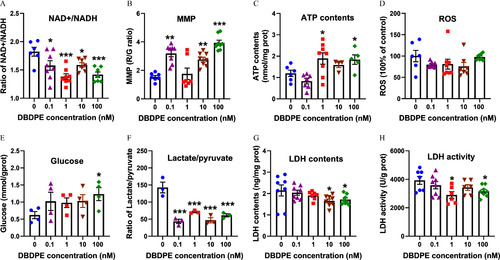
Energy metabolic levels in mouse spermatogonial GC-1 cells exposed to DBDPE and oxamate (LDH inhibitor) |