- Title
-
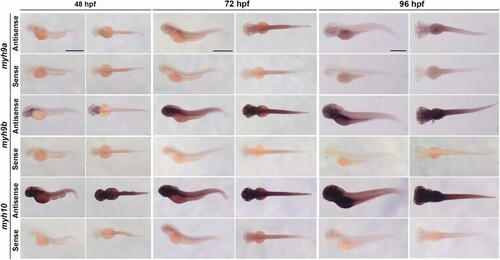
myh9b is a critical non-muscle myosin II encoding gene that interacts with myh9a and myh10 during zebrafish development in both compensatory and redundant pathways
- Authors
- Rolfs, L.A., Falat, E.J., Gutzman, J.H.
- Source
- Full text @ G3 (Bethesda)
|
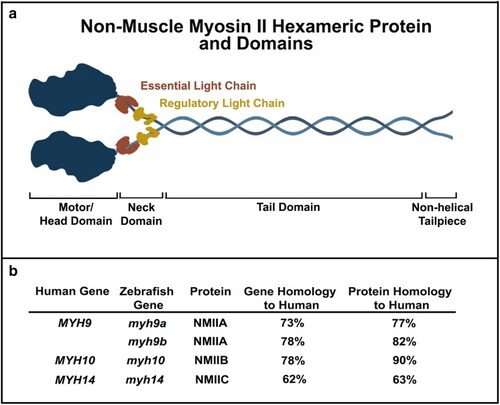
General NMII protein structure and homology. a) Schematic of the hexameric structure of an NMII protein. b) Zebrafish to human gene and protein homology. Zebrafish contain two |
|
|
|
|
|
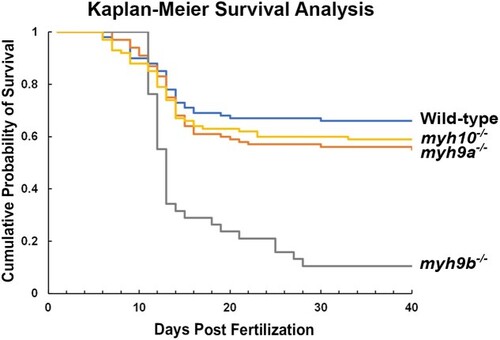
Kaplan–Meier survival curve for the PHENOTYPE:
|
|
|
|
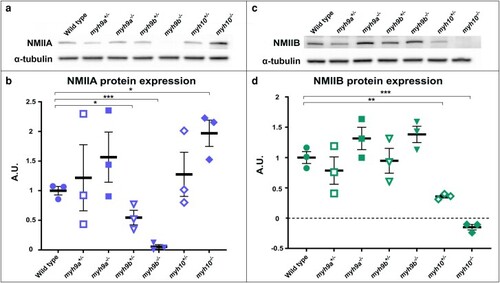
NMIIA and NMIIB protein levels in the zebrafish |
|
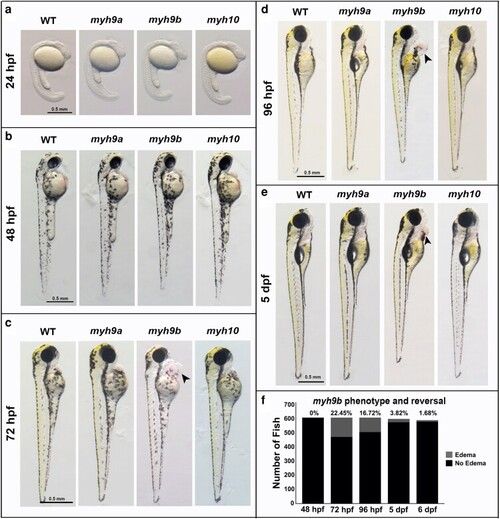
Double mutant phenotype quantification between 24 and 72 hpf and representative images at 72 hpf. a) Phenotype quantification for PHENOTYPE:
|
|
Summary of genetic interactions found through the analysis of the zebrafish |