- Title
-
Wnt/β-Catenin Signaling Regulates Yap/Taz Activity during Embryonic Development in Zebrafish
- Authors
- Astone, M., Tesoriero, C., Schiavone, M., Facchinello, N., Tiso, N., Argenton, F., Vettori, A.
- Source
- Full text @ Int. J. Mol. Sci.
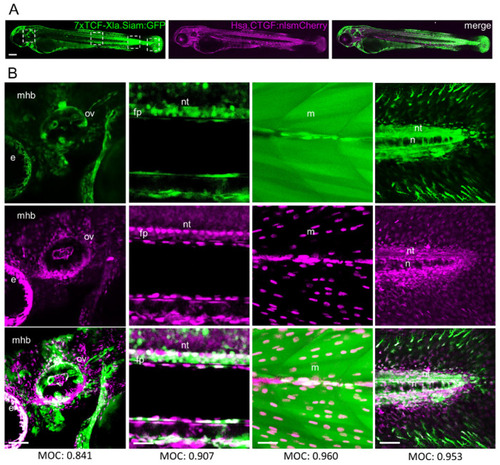
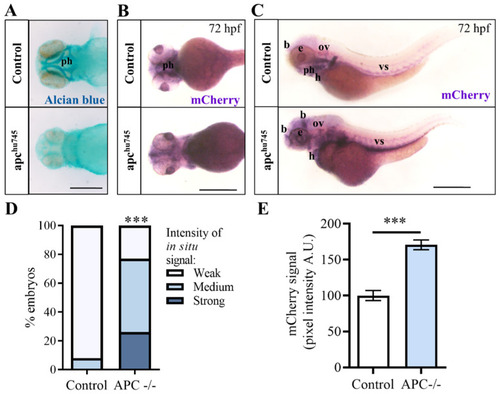
|
( |
|
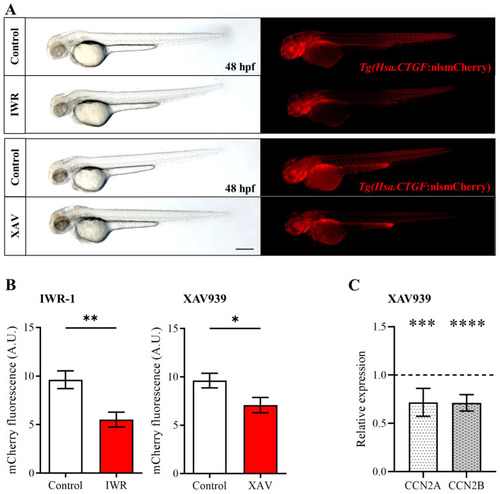
IWR-1 and XAV939-mediated β-catenin pathway inhibition reduces |
|
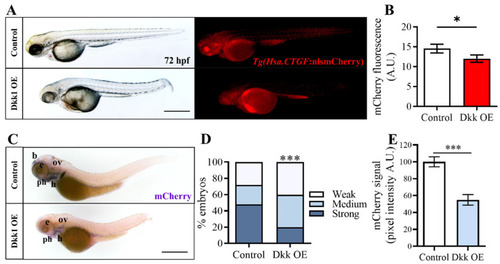
Wnt/β-catenin pathway inhibition through Dkk1 overexpression reduces |
|
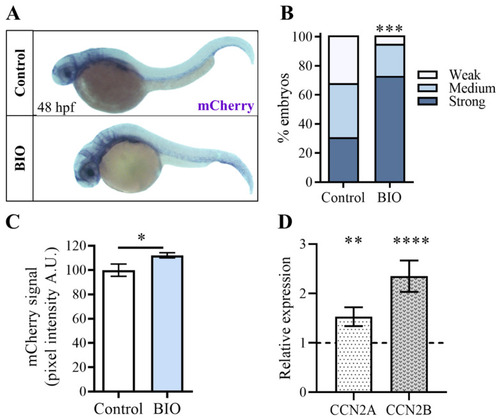
Pharmacological inhibition of β-catenin kinase GSK3 increases |
|
|