- Title
-
Beyond basic characterization and omics: Immunomodulatory roles of platelet-derived extracellular vesicles unveiled by functional testing
- Authors
- Palviainen, M., Puutio, J., Østergaard, R.H., Eble, J.A., Maaninka, K., Butt, U., Ndika, J., Kari, O.K., Kamali-Moghaddam, M., Kjaer-Sorensen, K., Oxvig, C., Aransay, A.M., Falcon-Perez, J.M., Federico, A., Greco, D., Laitinen, S., Hayashi, Y., Siljander, P.R.
- Source
- Full text @ J Extracell Vesicles
|
Interaction of macrophages with PEVs in vivo. Fluorescently labeled PEVs from platelets activated by TC co‐stimulation were intravenously injected into zebrafish embryos at 2 dpf and live‐imaged. (a) Left panel shows a simplified schematic of the site of microinjection and the main blood vessels including the ca, cv and cvp along with a representative BF image of the tissue area imaged. Right panel shows an overview of the experimental setup. (b)–(d) |
|
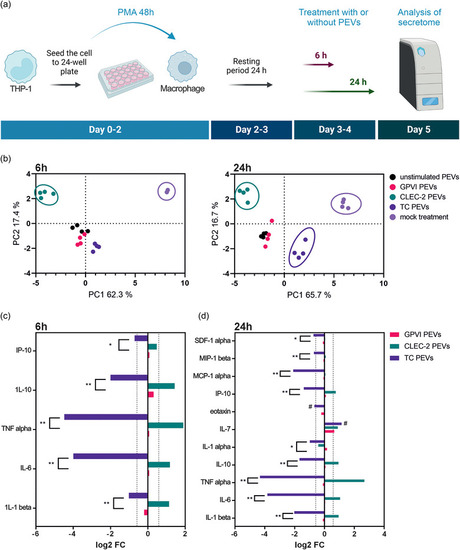
Changes in the macrophage cytokine secretomes induced by the different PEV types. (a) Flow chart of the experiment. THP‐1 cells were differentiated into macrophages by 48 h incubation with 50 nM PMA. After a resting period of 24 h, macrophages were treated with the different PEV types ( |
|
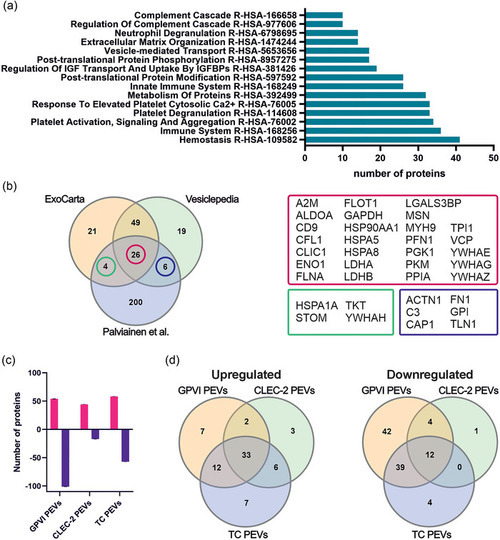
Comparison of the receptor‐induced differences in the protein cargo of PEVs analyzed with mass spectrometry ( |
|
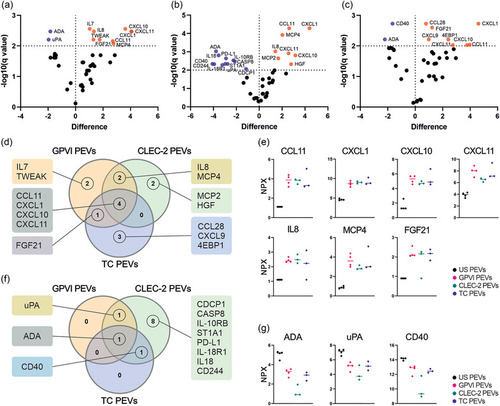
Quantitative analysis of the targets of the inflammation panel by PEA revealed distinctive receptor‐dependent tuning of the PEV proteins. PEVs isolated by iodixanol cushion ultracentrifugation were analysed for 80 inflammation panel targets. The GPVI ( |
|
Comparison of the miRNA content of unstimulated platelets and the differentially induced PEVs ( |