- Title
-
Functional analysis of ESRP1/2 gene variants and CTNND1 isoforms in orofacial cleft pathogenesis
- Authors
- Caetano da Silva, C., Macias Trevino, C., Mitchell, J., Murali, H., Tsimbal, C., Dalessandro, E., Carroll, S.H., Kochhar, S., Curtis, S.W., Cheng, C.H.E., Wang, F., Kutschera, E., Carstens, R.P., Xing, Y., Wang, K., Leslie, E.J., Liao, E.C.
- Source
- Full text @ Commun Biol
|
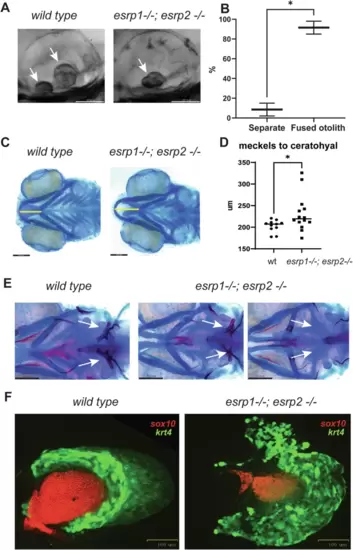
esrp1 and esrp2 are required for morphogenesis of epithelial-derived tissue: otoliths, pharyngeal teeth and pectoral fins.A zebrafish otoliths indicated by white arrows at 72 hpf. B Quantification and t-test of zebrafish otoliths from genotyped mutants characterized as separate or fused otoliths. t-test, n = 75. C Alcian blue representation of a 6 dpf zebrafish wildtype and esrp1−/− esrp2−/− double mutant showing cartilage stain, yellow line shows the measurement of the distance between the midline of Meckel’s and ceratohyal cartilages. D quantification and t-test analysis of this measurement in wildtype (n = 11) and esrp1−/−; esrp2 −/− mutants (n = 14). E Alcian blue and Alizarin red staining of larvae at 7 dpf ventral view, the pharyngeal teeth are present in wildtype (white arrows). In contrast, the esrp1−/−; esrp2−/− all exhibit decreased number of teeth, and occasionally some double mutants lack all ceratobranchial cartilages and the pharyngeal teeth are absent. F wildtype and esrp1−/− esrp2−/− mutant pectoral fins labeled with sox10 mCherry (red) and krt4 gfp (green). |
|
Complementary in vivo and in vitro functional assays to test human ESRP1 and ESRP2 gene variants.A Microdissected ANC of Alcian-blue stained embryos at 4 dpf for wildtype, esrp1−/−; esrp2+/+, esrp1−/−; esrp2−/−, and esrp1−/−; esrp2 MO embryos. B Schematic for the esrp morphant variant assay in zebrafish. Variants that robustly rescued the cleft ANC phenotype were scored as benign, while variants that failed to rescue the cleft ANC phenotype were scored as pathogenic. C RT-PCR was performed using primers spanning exons 36–38 of Arhgef11 on cDNA isolated from wildtype mouse Py2T cells, Esrp1/2 double-knockout Py2T cells, or Esrp1/2 double-knockout PyY2T cells electroporated with plasmids encoding for either Esrp1 or Esrp2 genes. Arrow markers point to the epithelial (short) isoform and mesenchymal (long) isoform retaining exon 37. PHENOTYPE:
|
|
Functional testing of human ESRP1 and ESRP2 gene variants.A Representative images of the ANC from Alcian-blue stained larvae at 4 dpf after injection with esrp2 MO and 200 pg of: esrp1 mRNA, esrp2 R353Q mRNA. ANC was scored as a rescued ANC or cleft ANC (B) ESRP1 and (C) ESRP2 gene variant rescue assay results for embryos injected with esrp2 MO and 200 pg of esrp1 variant mRNA. Results presented as percentage of rescue vs. cleft as different numbers of embryos survived and were analyzed, indicated as n above each bar. D ESRP1 and (E) ESRP2 gene variant rescue assay by detecting alternative splicing of Arhgef11 in murine Py2T wildtype and Esrp1−/−; Esrp2−/− double knockout cells. PHENOTYPE:
|
|
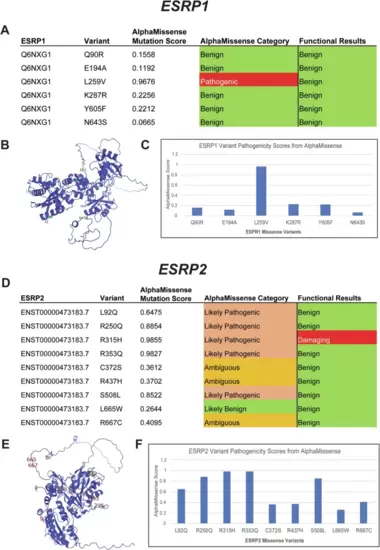
AlphaMissense pathogenicity predictions for ESRP1 and ESRP2 missense variants.ESRP1 and ESRP2 gene variants from OFC cases in the GMFK Children’s dataset and ClinVar variants associated with cleft lip and/or palate or autosomal recessive deafness were identified. Six ESRP1 and nine ESRP2 (A), (D) missense variants were analyzed using the AlphaMissense (AM) model. The tables (C), (F) show the AM-predicted pathogenicity compared to our functional test results and the AM mutation score, which is also graphed. On the left, the ESRP1 and ESRP2 AlphaFold structures (B), (E), with labeled missense mutations, color-coded with the functional results. |
|
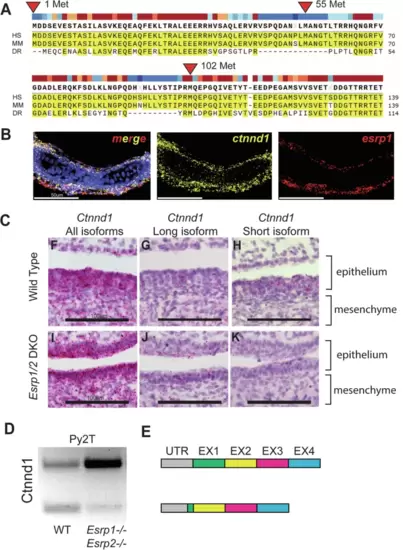
Alternative splicing of Ctnnd1 is regulated by Esrp1/2.A Amino acid sequence alignment of the first 140 residues of CTNND1 protein across human, mouse, and zebrafish. Translation for isoform 1 of CTNND1 begins at methionine 1, while isoform 3 encodes a truncated form that starts translation at methionine 102. Methionine residues at positions 55 and 324 are not conserved across all three species. B Detection of esrp1 and ctnnd1 gene expression in zebrafish at 4 dpf, demonstrates shared localization of transcripts in the embryonic epithelium. This coronal section includes the ventral Meckel’s cartilage. C Detection of murine Ctnnd1 mRNA using isoform-specific base-scope probes in the oral epithelium and tongue mesenchyme. The wildtype sections show that the Ctnnd1 long isoform is present in both epithelial and mesenchymal cells. The Ctnnd1 short isoform is present preferentially in epithelial cells and not in the mesenchymal cells. In the Esrp1/2 DKO mouse, the mesenchymal Ctnnd1 long isoform is detected in epithelial and mesenchymal cells, with loss of the Ctnnd1 short isoform. D RT-PCR of the Ctnnd1 long and short isoforms from Py2T cells. E Diagrammatic representation of the ESRP-regulated CTNND1 alternative splicing to generate the shorter epithelial isoform. EXPRESSION / LABELING:
|
|
Over-expression of ctnnd1 rescues esrp1−/−, esrp2−/− epithelial phenotypes.A Image representing how wildtype, intermediate and cleft ANC, pectoral fins and otoliths were sorted. B Representative table with the number of total fish injected and rescued by the ctnnd1 mRNA injection with GFP mRNA injection as control. Scoring of ANC phenotype (%) (C), fin phenotype (%) (D) and the otolith phenotype (%) (E) in the injected esrp1−/−; esrp2+/− inter-cross larvae confirmed by genotyping, showing 20–22% rescue of ANC, fin and otolith phenotypes in the esrp1−/−; esrp2−/− double homozygous larvae. PHENOTYPE:
|