- Title
-
ccdc141 is required for left-right axis development by regulating cilia formation in the Kupffer's vesicle of zebrafish
- Authors
- Wang, P., Shi, W., Liu, S., Shi, Y., Jiang, X., Li, F., Chen, S., Sun, K., Xu, R.
- Source
- Full text @ J. Genet. Genomics
|
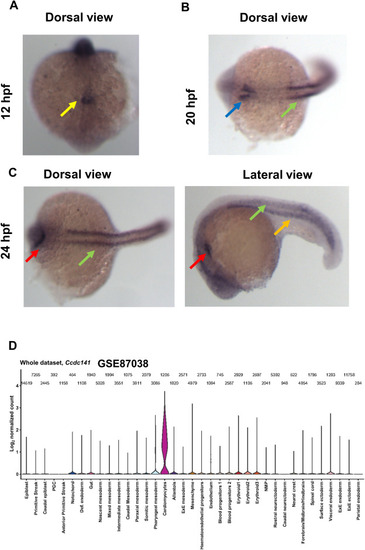
Distribution of ccdc141/Ccdc141 expression in zebrafish and mice. A–C: Results of whole mount in situ hybridization of ccdc141 in zebrafish, showing expression in the KV (yellow arrow), otic vesicle (blue arrow), somites (green arrows), heart tube (red arrows), and intermediate mesoderm (orange arrow). D: The expression of Ccdc141 in mouse gastrulation and early organogenesis according to GSE87038 is shown. Violins are width-normalized. The numbers above each violin show the number of cells in the group. KV, Kupffer’s vesicle; hpf, hours post fertilization. |
|
ccdc141 crispants in zebrafish displayed abnormal LR patterning. A: The schematic diagram documenting the process and guide RNAs used for generating ccdc141 crispants. B: Cardiac looping of zebrafish at 48 hpf is presented in the Tg(myl7:RFP) line in the ventral view. Normal D-loop, abnormal N-loop, and L-loop are shown. C: The quantification of cardiac looping (percentage). Cas9 control/crispants/crispants + mRNA, N = 3; n = 164/213/196; ∗∗∗∗, P < 0.0001; two-sided Fisher's exact test for crispants with control and crispants + mRNA with control. D: Double WISH for myl7 (heart) and ins (pancreas) at 72 hpf. SA indicates any intermediate state between SS and SI. E: The percentage of abnormal organ situs with crispants. Control vs. crispants, N = 3; n = 63/107. ∗∗∗∗, P < 0.0001; two-sided Fisher's exact test for crispants with control. F: Representative images of normal zebrafish (above), pericardial edema (middle), and curved tail (bottom) at 48 hpf. G and H: pericardial edema and curved tail of zebrafish (percentage). N = 6; ∗∗∗∗, P < 0.0001; ∗∗, P < 0.01; ∗, P < 0.05; ns, not significant; two-sided Fisher's exact. WT, wild-type; R, right; L, left; SA, situs ambiguous; SS, situs solitus; SI, situs inversus; hpf, hours post fertilization. |
|
ccdc141 exhibited global effects on early signaling pathways in LR development. A–C: Expression patterns of spaw (A, 16 hpf), lefty2 (B, 17 hpf), and pitx2 (C, 18 hpf) in zebrafish embryos. Left (normal), right (abnormal), bilateral (abnormal), or absent (abnormal). D–F: The quantification of the gene expression. Cas9 vs. crispants, N = 3; n = 91/67 (D); n = 91/67 (E); n = 91/67 (F). ∗∗, P < 0.01; ∗∗∗∗, P < 0.0001; two sided Fisher's exact test. L, left; R, right. |
|
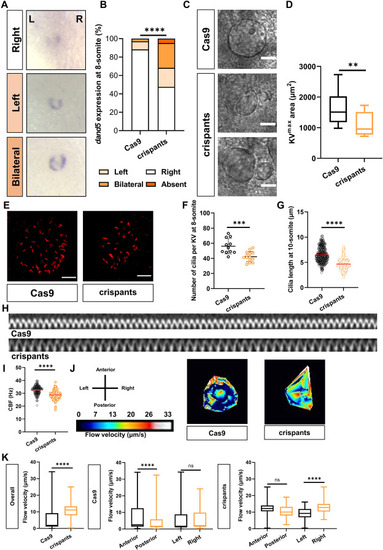
ccdc141 is required for asymmetry decay of dand5 and normal KV flow. A and B: The correct expression (right) and aberrant expression (left, bilateral) of dand5 at 8 ss are shown. Control vs. crispants, N = 3; n = 34/63; ∗∗∗∗, P < 0.0001; two-sided Fisher's exact test. C and D: Representative DIC images (C) and qualification (D) of different KVs in control and crispants embryos at 8 ss. Control vs. crispants, n = 30/10; ∗∗, P < 0.01. E–G: Knockdown of ccdc141 decreases the cilia number per KV, shorter the cilia length. Cilia number, n = 11/14 embryos. ∗∗∗, P < 0.001; two-sided unpaired t-test. Cilia length, N = 11/14; n (cilia) = 293/282. ∗∗∗∗, P < 0.0001; Mann-Whiteney test. H and I: The kymograph and the quantification of cilia beating. Control vs. crispants, N = 9/10, n (cilia) = 158/124. ∗∗∗∗, P < 0.0001; Mann-Whiteney test. J: Heat map of flow speed showing detailed regions within the KV in control and crispant embryos at the 8 ss. The pseudo-color scale represents flow speed in μm/s, where white represents high speed versus low speed in blue. K: Box plots for instantaneous flow speed measured at different locations of the KVs, based on the same data set used to generate the heat maps. Box plots display the median with a vertical line, and the whiskers represent the minimum and maximum values observed. Control vs. crispants, Ne, number of embryos; ntracks, number of particle tracks followed. Ne = 2/2, overall ntracks = 599/168, anterior ntracks = 235/65, posterior ntracks = 364/103, left ntracks = 358/86, right ntracks = 241/82. ∗∗∗∗, P < 0.0001; Mann-Whiteney test. Scale bars, 30 μm (C); 20 μm (E). L, left; R, right; KV, Kupffer’s vesicle. |
|
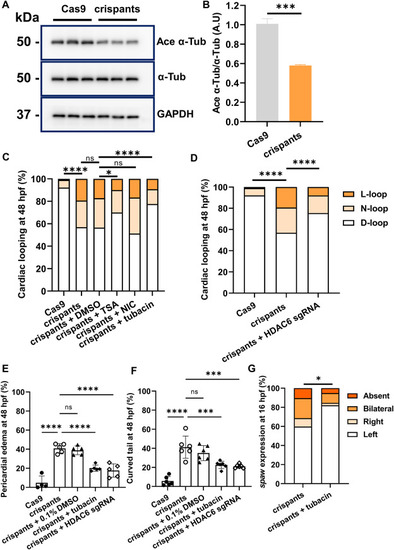
hdac6 can rescue the ciliopathy-like phenotype of ccdc141 crispants. A: Protein expression of acetylated-α-tubulin (Ace-α-Tub) and α-tubulin (α-Tub) in zebrafishes of the control and ccdc141 crispants group was evaluated by Western blot. GAPDH served as a loading control. B: Quantification of acetylated-α-tubulin protein level when normalized against α-tubulin. N = 3; ∗∗∗, P < 0.001; Student's t-test. A.U, arbitrary units. C: The quantification of cardiac looping in zebrafish treated by DMSO (control) or different pharmaceuticals. Trichostatin A (TSA), a pan-HDAC inhibitor; Nicotinamide (NIC), an inhibitor of sirt1 and sirt2; Tubacin, a highly selective HDAC6 inhibitor. N = 3; n = 206/244/256/100/78/228. ∗∗∗∗, P < 0.0001; ∗, P < 0.05; ns, P > 0.05; one-way ANOVA test. D–F: The abnormal cardiac looping, pericardial edema, and curved tail in the ccdc141 crispants embryos under the inhibition of hdac6. N = 3; n = 206/244/266; ∗∗∗∗, P < 0.0001; Fisher's exact test (D). N = 5/6; ∗∗∗∗, P < 0.0001; ∗, P < 0.05; ns, P > 0.05; one-way ANOVA test (E and F). G: Tubacin was able to partially rescue the spaw expression pattern in ccdc141 crispants. n = 67/39; ∗, P < 0.05; two sided Fisher's exact test. |
|
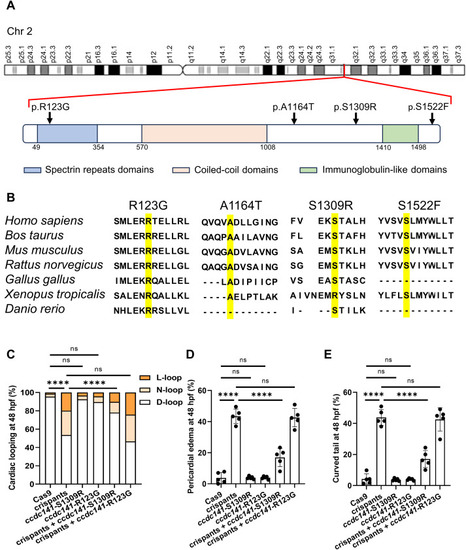
The damaging CCDC141 rare variant in patients with laterality defects. A: The genomic location and the amino acid changes of the four CCDC141 variants. B: CCDC141 proteins alignment highlighting the conservation of the affected amino acids in patients with laterality defects (yellow). C–E: The abnormal cardiac looping, pericardial edema, and curved tail in the ccdc141 crispants embryos. N = 3; n = 201/293/128/257 (C). N = 5 (D and E). ∗∗∗∗, P < 0.0001; ns, no significance; two-sided Fisher's exact test. |
Reprinted from Journal of genetics and genomics = Yi chuan xue bao, 51(9), Wang, P., Shi, W., Liu, S., Shi, Y., Jiang, X., Li, F., Chen, S., Sun, K., Xu, R., ccdc141 is required for left-right axis development by regulating cilia formation in the Kupffer's vesicle of zebrafish, 934-946, Copyright (2024) with permission from Elsevier. Full text @ J. Genet. Genomics