- Title
-
Chemical screen in zebrafish lateral line identified compounds that ameliorate neomycin-induced ototoxicity by inhibiting ferroptosis pathway
- Authors
- Fan, Y., Zhang, Y., Qin, D., Shu, X.
- Source
- Full text @ Cell Biosci.
|
Otoprotection against various ototoxin-induced lateral line neuromast damage by inhibitors of different cell death pathways. Typical confocal images and quantitative results of otoprotection assay. ( |
|
ROS staining in hair cells. Representative confocal images of DCF staining ( |
|
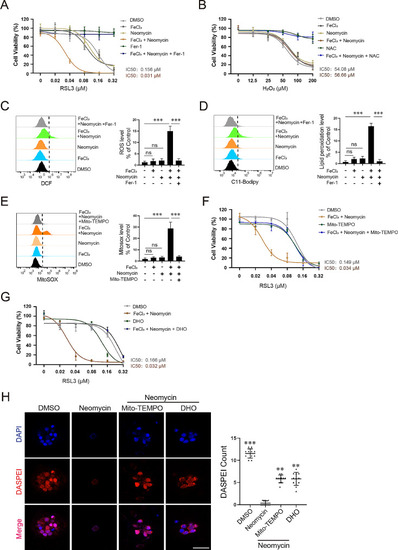
Neomycin and Fe3+synergistically sensitize HT1080 cells to ferroptosis inducer RSL3. ( |
|
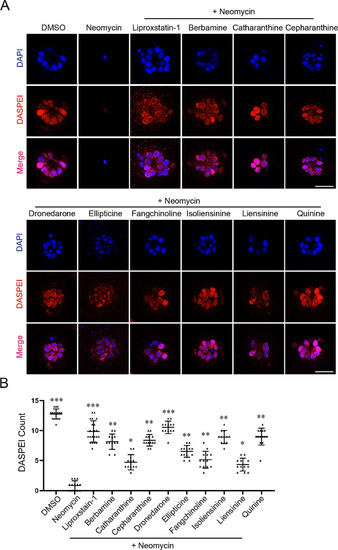
Otoprotection screening against neomycin-induced hair cell damage in zebrafish lateral line. A Representative otoprotection results of the nine positive hit compounds identified from our screen. Liperoxstatin-1 is a positive control. Scale bar: 20 μm. ( |
|
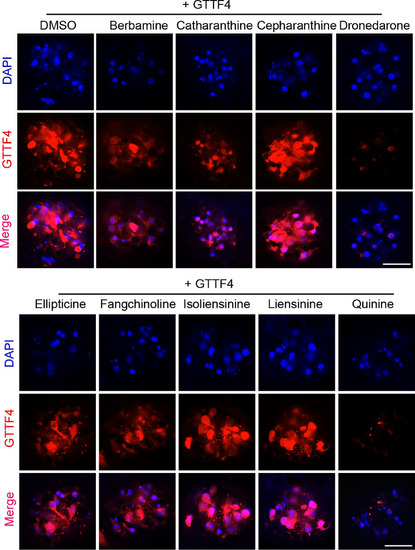
Dronedarone and quinine block the uptake of GTTF4. GTTF4 loading assay were performed to evaluate the ability of a testing compound to block the uptake of GTTF4 in hair cells. Assays were repeated twice, and representative images are shown here. Scale bar: 20 μm |
|
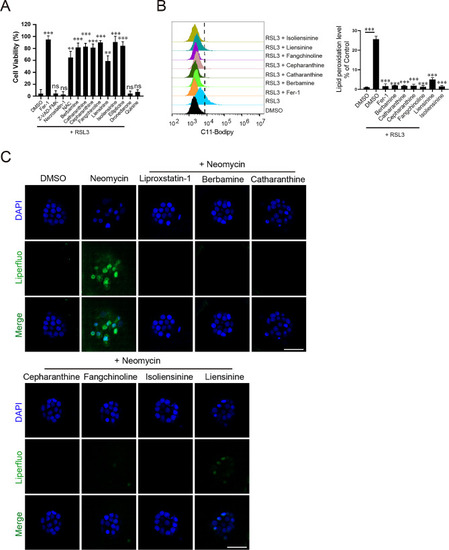
Multiple hit compounds inhibit lipid ROS accumulation and ferroptosis. A All hit compounds other than dronedarone and quinine are able to inhibit RSL3-induced ferroptosis in HT1080 cells. Data represent mean ± s.d. from three biological repeats and |
|
Ellipticine inhibits ferroptosis by downregulation of TFRC and iron homeostasis. A Ellipticine toxicity assay in HT1080 cells. Data represent mean ± s.d. from three biological repeats. ( |