- Title
-
A zebrafish xenotransplant model of anaplastic thyroid cancer to study the tumor microenvironment and innate immune cell interactions in vivo
- Authors
- Michael, C., Mendonça-Gomes, J.M., DePaolo, C.W., Di Cristofano, A., De Oliveira, S.
- Source
- Full text @ Endocr. Relat. Cancer
|
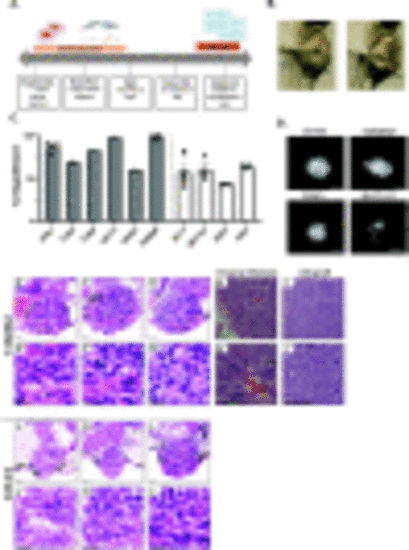
ATC cell lines engraft in zebrafish larvae at different rates. (A) Experimental timeline for thyroid xenotransplant model. (B) Representative images acquired using the stereomicroscope at 2 dpf to show proper site of injection. (C) Graph showing percent engraftment for ATC mouse (N794, T1903, T1860, N2773, N2933, T4888M) and human (ACT1, BHT101, ASH3, C643) cell lines. (D) Representative images acquired using the fluorescent microscope at 4 dpi in mouse and human ATC cell lines. Scale bar= 100 μm (E-G & JL) Representative images of T4888 and C643 ATC cell lines in zebrafish. Scale bar= 20 μm. (E’-G’ & J’-L’) Higher magnification showcasing important histopathological features maintained in T4888 and C643 ATC zebrafish xenotransplants. Scale bar= 20 μm. (H & H’) Histopathological analysis from original lung metastasis of T4888 ATC cells in a murine mouse model ((Pten, P53)/−/−). Scale bar= 50 μm. (I & I’) Histopathological analysis from an allograft in a murine mouse model. Scale bar= 50 μm. Data is from at least two independent experimental replicates. Bar plots show mean ± SEM, each dot represents one experiment. |
|
T4888M and C643 ATC cell lines display different growth and proliferation rates. (A) Graph showing mass volume μm3 at 1 dpi compared to 4 dpi in ATC cell lines T4888 and C643 (T4888 n=26, C643 n=27). (B) Graph showing mass volume μm3 at 1 dpi compared to 4 dpi tracking individual larva in ATC cell lines T4888 and C643. (C-D) Representative 3D images and reconstruction of ATC tumor masses from 1 dpi to 4 dpi in ATC cell lines T4888 and C643. Images were acquired using the 20x objective. Scale bar= 50 μm. (E) Representative 3D images of EDU+ cells inside cancer mass of ATC cell lines T4888 and C643. (F) Quantification of the total number of EDU+ cells in T4888 and C643 ATC masses (T4888 n=15, C643 n=16). Images were acquired using the 40x objective. Scale bar= 30 μm. (G) Quantification of the number of EDU+ cells normalized by the area μm2 of the ATC mass. Data is from at least two independent experimental replicates. Bar plots show mean ± SEM, each dot represents one larva, P-values are shown in each graph. |
|
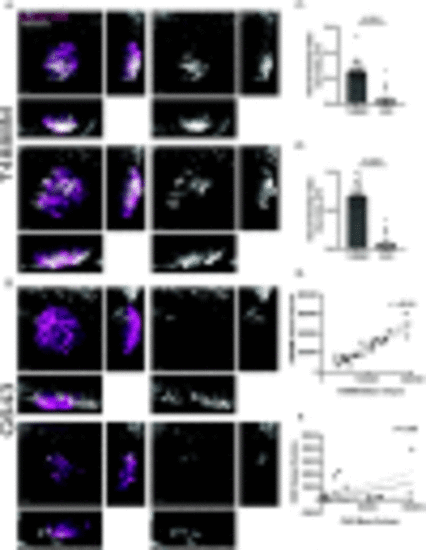
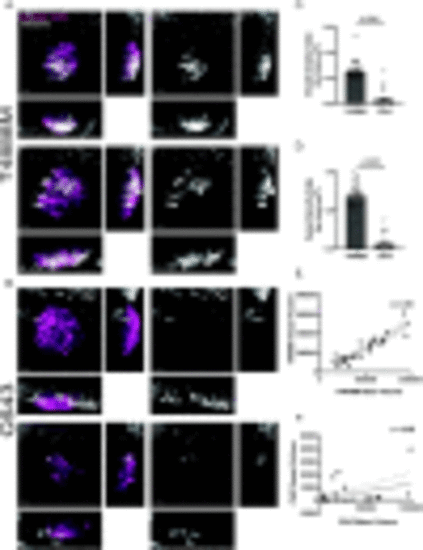
T4888M ATC xenotransplants display high angiogenic ability. (A-B) Representative 3D images of ATC tumor mass and vessel recruitment at 4 dpi Tg(fli:GFP) in ATC cell lines T4888 and C643. Scale bar= 50 μm. (C) Graph showing vessel density in tumor mass normalized by tumor volume μm3 for ATC cell lines T4888 and C643 (T4888 n=26, C643 n=26). (D) Graph showing vessel density in tumor mass normalized by tumor area μm2 for ATC cell lines T4888 and C643 (T4888 n=26, C643 n=26). (E) Graph of the correlation between T4888 mass volume and T4888 vessel volume. (F) Graph of the correlation between C643 mass volume and C643 vessel volume (T4888 n=26, C643 n=26). Data is from at least two independent experimental replicates. Bar plots show mean ± SEM, each dot represents one larva, P-values are shown in each graph. Pearson correlation graphs show line of best fit; each dot represents one larva, r-values are shown in each graph. |
|
In vivo visualization of T4888M cell cycle using the PIP-FUCCI reporter. (A) Representative 3D images of T4888 ATC cancer cells using the PIP-FUCCI reporter. Scale bar= 30 μm. (B) Graph showing total number of cells at 4 dpi in each cell cycle phase with the PIP-FUCCI reporter (T4888 n=22). (C) Graph showing cell cycle phases with the PIP-FUCCI reporter normalized by the total number of cells in T4888 ATC mass (T4888 n=22). (D-E) Images from 48-hour live imaging to highlight proliferative cells, imaging begins 1dpi and ends at 3dpi. Data is from at least two independent experimental replicates. Bar plots show mean ± SEM, each dot represents one larva, P-values are shown in each graph. |
|
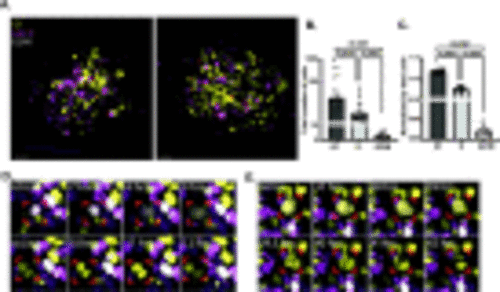
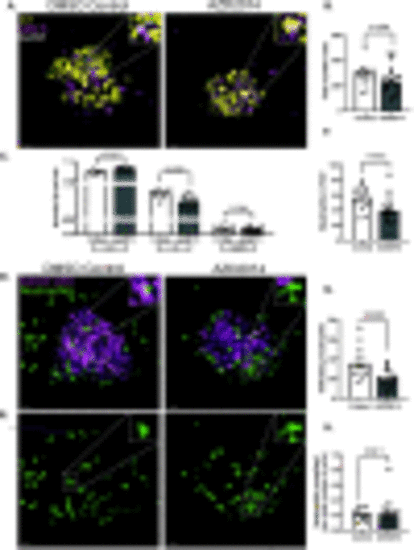
T4888M ATC cell line responds to cell cycle inhibitors. (A-C) Representative 3D images of ATC tumor mass and neutrophil recruitment at 4 dpi Tg(lyzC:BFP) with DMSO control and AZD2014 treatment. Zebrafish larvae were treated with 0/1% DMSO control or 1 μm AZD2014 (Vistusertib). Scale bar= 20 μm. (D) Graph showing cell cycle phases with the PIP-FUCCI reporter normalized by the total number of cells in DMSO control and AZD2014 treatment (DMSO Control n=14, AZD2014 n=22). (E) Graph showing total number of cells in each mass in DMSO control and AZD2014 treatment (DMSO Control n=14, AZD2014 n=22). (F) Graph showing the total number of neutrophils in field of view (FOV) for the DMSO control and AZD2014 (DMSO Control n=14, AZD2014 n=18). (G) Graph showing total number of neutrophils infiltrating the mass in the DMSO control and AZD2014 (DMSO Control n=14, AZD2014 n=18). (H) Graph showing the total number neutrophils normalized by the total number of cells in each larva (DMSO Control n=14, AZD2014 n=18). Data is from at least two independent experimental replicates. Bar plots show mean ± SEM, each dot represents one larva, P-values are shown in each graph. |