- Title
-
col1a2+ fibroblasts/muscle progenitors finetune xanthophore countershading by differentially expressing csf1a/1b in embryonic zebrafish
- Authors
- Chen, J., Wang, H., Wu, S., Zhang, A., Qiu, Z., Huang, P., Qu, J.Y., Xu, J.
- Source
- Full text @ Sci Adv
|
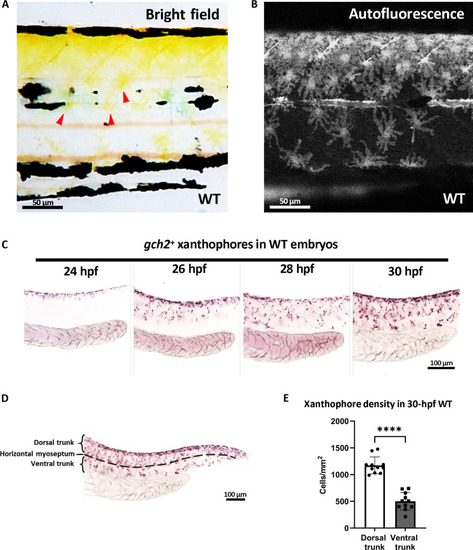
Xanthophore countershading in embryonic zebrafish. ( |
|
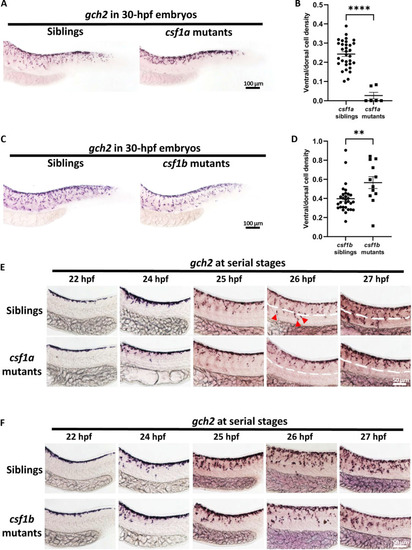
Xanthophore countershading is abnormal in ( |
|
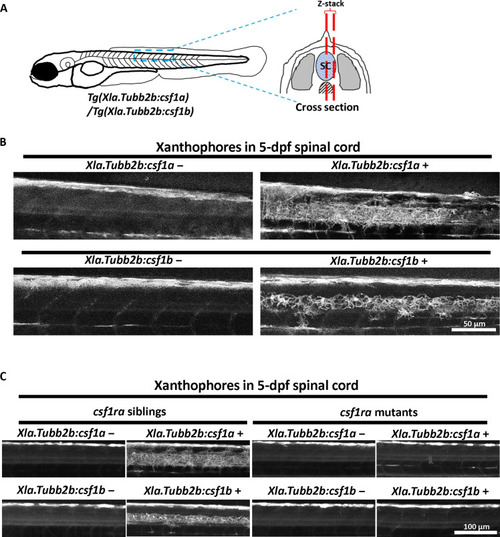
Csf1a and Csf1b are chemoattractants of xanthophores. ( |
|
( |
|
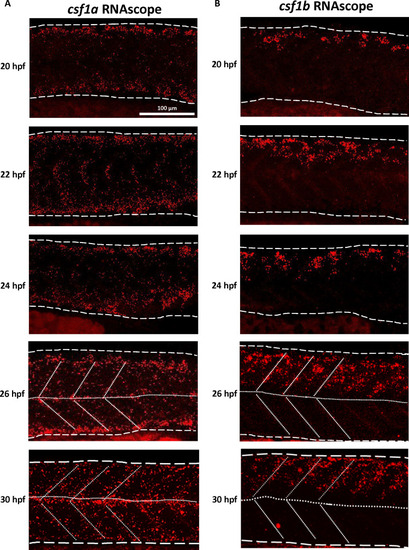
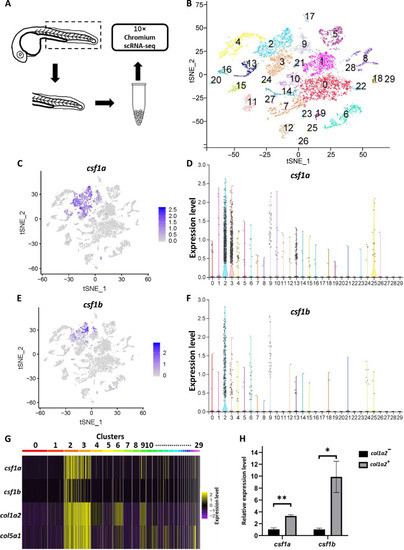
scRNA-seq reveals the sources of Csf1a and Csf1b in embryonic zebrafish. ( |
|
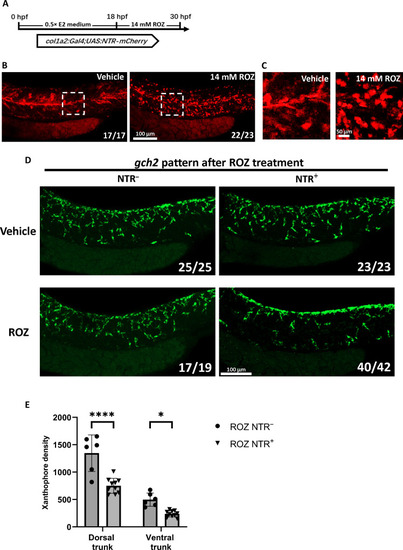
Ablation of ( |