- Title
-
Rab11 promotes single Mauthner cell axon regeneration in vivo through axon guidance molecule Ntng2b
- Authors
- Yao, H., Shen, Y., Song, Z., Han, A., Chen, X., Zhang, Y., Hu, B.
- Source
- Full text @ Exp. Neurol.
|
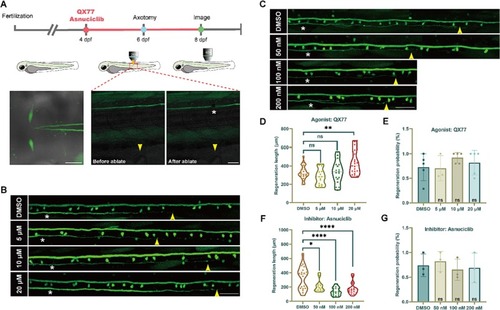
Systemic intervention with Rab11 promoted M-cells axon regeneration. (A) Above is timeline of time points of pharmacological treatment, axotomy and imaging. Below are diagram of M-cells soma and crossed axon initial segment. Representative images of the M-cell axon before and after ablation by two-photon laser. Asterisk, ablation site; Arrowhead, cloacal pore; Scale bar, 50 μm. (B, C) Representative diagram of confocal imaging of M-cells axon regeneration between DMSO and agonist/inhibitor (concentration gradient, QX77: 5 μΜ, 10 μM, 20 μM; Asnuciclib: 50 nM, 100 nM, 200 nM). Asterisk, ablation site; Arrowhead, axon regeneration terminal; Scale bar, 50 μm. (D, F) Violin plot shows that statistical quantification of axon regeneration. (QX77: DMSO, 326.9 ± 14.89 μm, n = 24; 5 μΜ, 279.4 ± 22.19 μm, n = 15; 10 μM, 333.8 ± 28.12 μm, n = 18; 20 μM, 433.0 ± 26.67 μm, n = 21. Asnuciclib: DMSO, 328.7 ± 37.14 μm, n = 19; 50 nM, 211.8 ± 23.07 μm, n = 13; 100 nM, 136.6 ± 12.79 μm, n = 18; 200 nM, 179.3 ± 20.09 μm, n = 18.) Assessed by one-way ANOVA. * p < 0.05, ** p < 0.01, **** p < 0.0001, ns, no significance. (E, G) Statistical diagram of successful probability of M-cells axon regeneration after drugs treatment with different concentrations (probability range: 70% - 92%. QX77: DMSO, 72.6 ± 12.2%; 5 μΜ, 70.5 ± 14.8%; 10 μM, 92 ± 4.9%; 20 μΜ, 82 ± 11.4%; Asnuciclib: DMSO, 74.1 ± 13.3%; 50 nM, 81.39 ± 11.7%; 100 nM, 65.2 ± 12.5%; 200 nM, 68.9 ± 17.36%). The results were obtained from 3 to 5 independent experiments. Assessed by unpaired, two-tailed Student's t-test. ns, no significance. |
|
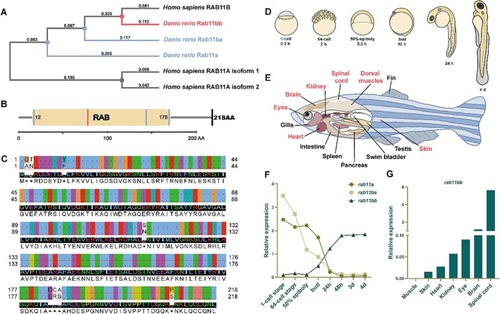
Expression patterns of different Rab11 isoforms in zebrafish. (A) The phylogenetic tree with the maximum-likelihood algorithm. The number above the evolutionary branch represents the distance of homology. (B) Protein functional domain of RAB11B (Homo sapiens) and Rab11bb (Danio rerio). (C) Sequence alignment between RAB11B (Homo sapiens) and Rab11bb (Danio rerio). Above is RAB11B and below is Rab11bb. (D) The schematic diagram of developmental stages of zebrafish embryo after fertilization. (E) Anatomical diagram of adult zebrafish (red marked: tissues for subsequent experiment). (F) Line graph of qRT-PCR analysis of different Rab11 isoforms in zebrafish larvae at different developmental stages. The total RNA of each stage was obtained from 30 zebrafish larvae by three independent experiments. (G) Histogram of relative expression analysis of Rab11bb in adult zebrafish at different tissues. The total RNA of each tissue was obtained from 4 adult zebrafish by three independent experiments. |
|
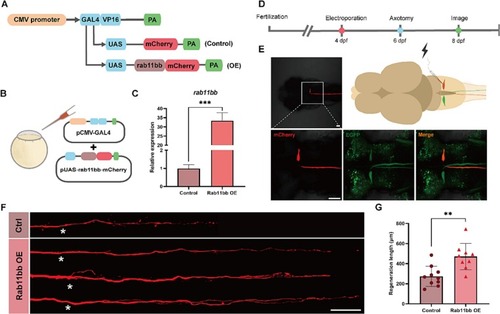
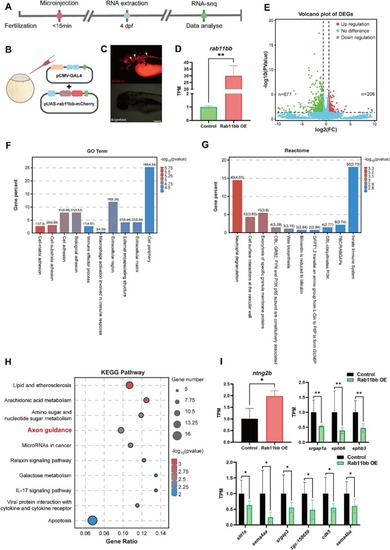
Rab11bb overexpression promoted the capacity of M-cells axon regeneration. (A) Plasmid construction legend of the control group and Rab11bb overexpression plasmids. The CMV-GAL4-VP16 plasmid drives the expression of UAS-mCherry and UAS-rab11bb-mCherry plasmids (CMV: Cytomegalovirus; GAL4: Galectin-4; PA: poly adenylate). (B) Schematic diagram of microinjection using the two-plasmid system. (C) qRT-PCR analysis of rab11bb overexpression efficiency. The total RNA of each group (n = 30) was obtained from three independent experiments. Assessed by unpaired, two-tailed Student's t-test. *** p < 0.001. (D) Timeline of time points of electroporation, axotomy and imaging. (E) Pattern Diagram of electroporation and confocal images of positive expression in M-cell. The white box represents the position of M-cell under 40 × magnification in 10 × magnification field. Scale bar, 50 μm. (EGFP: labeled M-cells in Tol-056 zebrafish strain; mCherry: fluorescent reporter gene in foreign plasmid). (F) Representative diagram of confocal imaging of M-cells axon regeneration. Asterisk, ablation site; Scale bar, 50 μm. (Ctrl: control; OE: overexpression). (G) Statistical quantitative diagram of axon regeneration (control: 273.0 ± 31.71 μm, n = 10; Rab11bb overexpression: 470.5 ± 43.82 μm, n = 9). Assessed by unpaired, two-tailed Student's t-test. ** p < 0.01. |
|
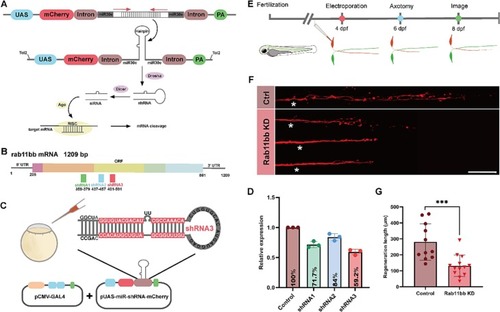
Rab11bb knockdown suppressed the capacity of M-cells axon regeneration. (A) Schematic diagram of working principle of primary miR-shRNA gene silencing system (RISC: RNA-induced silencing complex). (B) The position of shRNA1–3 in open reading frame (ORF) of rab11bb (UTR: untranslated region). (C) Schematic diagram of microinjection using the miR-shRNA system. (D) qRT-PCR analysis of rab11bb knockdown efficiency. The total RNA of each group (n = 30) was obtained from three independent experiments (shRNA1: 71.7 ± 3.0%; shRNA2: 84.0 ± 3.4%; shRNA3: 59.2 ± 2.7%). (E) Timeline of time points of electroporation, axotomy and imaging. (F) Representative diagram of confocal imaging of M-cells axon regeneration. Asterisk, ablation site; Scale bar, 50 μm (Ctrl: control; KD: knockdown). (G) Statistical quantitative diagram of axon regeneration (control: 280.6 ± 34.48 μm, n = 11; Rab11bb knockdown: 131.0 ± 18.63 μm, n = 13). Assessed by unpaired, two-tailed Student's t-test. *** p < 0.001. |
|
RNA-seq revealed that the regenerative promoting mechanism of Rab11bb was related to the axon guidance pathway. (A) Timeline of time points of microinjection, RNA extraction and RNA-seq. (B, C) Schematic diagram of microinjection to overexpress Rab11bb and the positive expression of overexpressed plasmid at 4 dpf. Scale bar, 0.2 mm. (D) RNA-seq analysis of rab11bb overexpression efficiency. The total RNA of each group (n = 30) was obtained from three independent experiments. Assessed by unpaired, two-tailed Student's t-test. ** p < 0.01. (E) The volcano plot shows the expression profiling between the two groups. The vertical dotted lines refer to a 2.0-fold (log2 scaled) up-regulation and down-regulation, respectively. The horizontal dotted line corresponds to a p-value of 0.05 (−log10 scaled). The red and green points in the plot represent DEGs with statistical significance. * p < 0.05. (F) The GO enrichment analysis (biological process terms) of DEGs. The figure shows top 10 terms on the x-axis that are significantly enriched by a p-value of 0.05 (−log10 scaled). The y-axis displays the gene percent to all GO biological process terms. (G) The Reactome pathway enrichment analysis of DEGs. The figure shows top 10 pathways on the x-axis that are significantly enriched by a p-value of 0.05 (−log10 scaled). The y-axis displays the gene percent to all Reactome pathways. (H) Enrichment for KEGG pathway analysis of DEGs. The x-axis displays the gene ratio in each pathway. The y-axis shows the top 10 pathways that are significantly enriched. The bubble size indicates the number of genes. The color bar indicates the corrected p-value, the red represents higher value, the blue represents lower value. p-value of 0.05 (−log10 scaled). (I) Quantitative histogram of expression analysis of the DEGs in the axon guidance pathway by RNA-seq between experiment and control group. * p < 0.05, ** p < 0.01. |
|
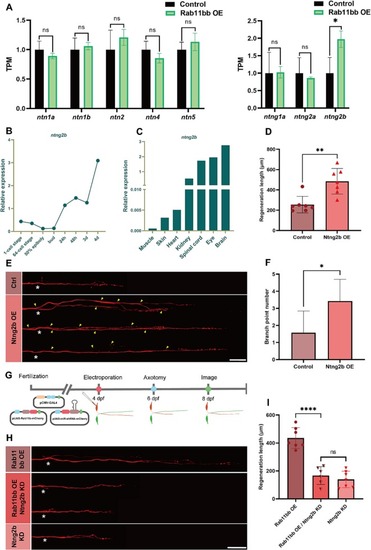
The expression of ntng2b was analyzed and overexpression promoted M-cells axon regeneration. (A) Quantitative histogram of netrin family expression analysis by RNA-seq between experiment and control group. Assessed by unpaired, two-tailed Student's t-test. * p < 0.05, ns, no significance. (B) Line graph of relative expression analysis of ntng2b in zebrafish larvae by qRT-PCR at different developmental stages. The total RNA of each stage was obtained from 30 zebrafish larvae by three independent experiments. (C) Histogram of relative expression analysis of ntng2b by qRT-PCR in adult zebrafish at different tissues. The total RNA of each tissue was obtained from 4 adult zebrafish by three independent experiments. (D) Statistical quantitative diagram of axon regeneration (control: 254.9 ± 30.71 μm, n = 7; Ntng2b overexpression: 484.8 ± 47.62 μm, n = 7). Assessed by unpaired, two-tailed Student's t-test. ** p < 0.01. (E) Representative diagram of confocal imaging of M-cells regenerated axons and branches. Asterisk, ablation site; Arrowhead, axon branch points; Scale bar, 50 μm. (F) Statistical quantitative diagram of axon branch point number (control: 1.57 ± 0.48, n = 7, total branch points number = 11; Ntng2b overexpression: 3.43 ± 0.48, n = 7, total branch points number = 24). Assessed by unpaired, two-tailed Student's t-test. * p < 0.05. (G) Timeline of time points of electroporation, axotomy and imaging. (H) Representative diagram of confocal imaging of M-cells regenerated axons. Asterisk, ablation site; Scale bar, 50 μm. (I) Statistical quantitative diagram of axon regeneration (Rab11 overexpression: 435.7 ± 28.23 μm, n = 7, Rab11bb overexpression / Ntng2b knockdown: 166.6 ± 25.97 μm, n = 6, Ntng2b knockdown: 139.1 ± 22.25 μm, n = 7). Assessed by unpaired, two-tailed Student's t-test. **** p < 0.0001, ns, no significance. |